Inspiration
Our project was inspired by the principles of natural selection and evolution, where traits are optimized over generations to maximize fitness. In the context of CRISPR, one of the greatest risks lies in unintended off-target effects, which can have harmful consequences in therapeutic or research settings. We drew on the analogy of evolution to design a system that uses natural selection as a framework to minimize off-target effects while maximizing on-target precision, mirroring the way biological systems refine traits for survival.
What it does
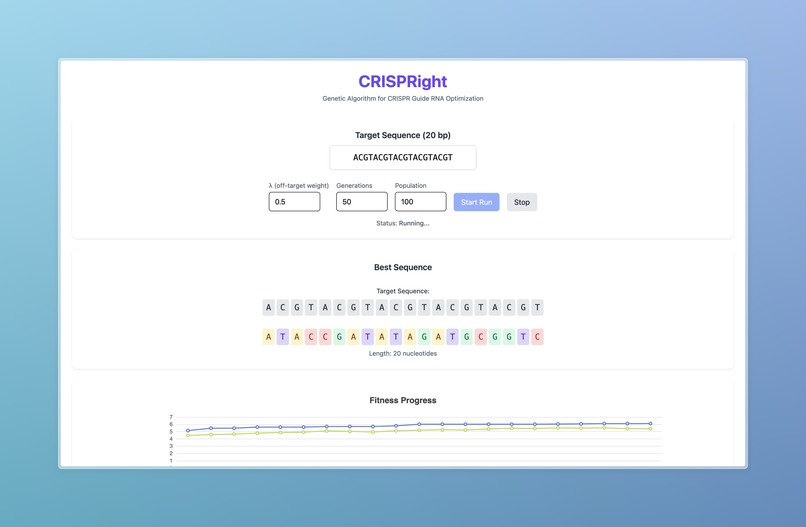
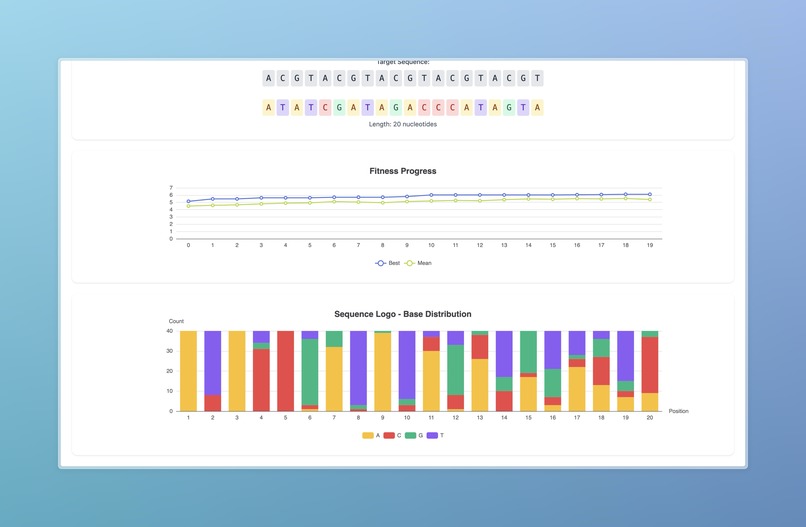
The project simulates natural selection to engineer the most optimal guide RNA for a given target sequence. By modeling populations of candidate sequences and applying evolutionary pressures, it demonstrates how iterative selection can optimize precision. This not only highlights the parallels between evolution and computational optimization but also produces a workflow that generates interpretable, biologically meaningful insights into guide RNA engineering.
How we built it
We began by cleaning and standardizing data, producing consistent off-target risk metrics using both CFD and CRISPRoff scores. We then combined a genetic algorithm with a sequence embedding model (DNA-BERT), feeding into a multilayer perceptron to evaluate candidate guide RNAs. The system simulates generations of populations, applying genetic heuristics such as mutation and crossover to produce biologically plausible and functional guide RNAs. To evaluate each new generation, we incorporated Cas-OFFinder to generate off-target predictions, providing a feedback loop that drives fitness and optimization across simulated evolutionary cycles.
Challenges we ran into
One of the main challenges was standardizing the heterogeneous data available for CRISPR off-target predictions. Extracting meaningful features from nucleotide sequences and translating them into usable inputs for the model also proved difficult. In addition, training the model within hackathon time constraints required balancing complexity with efficiency. Finally, replicating realistic evolutionary dynamics in a computational simulation required careful design of heuristics to balance biological plausibility with computational feasibility.
Accomplishments that we're proud of
We successfully developed a fully functional workflow that integrates three models to create visualized simulations of natural selection applied to guide RNA design. This workflow not only demonstrated the technical feasibility of the approach but also provided an interpretable and engaging way to understand the evolutionary optimization of CRISPR guide RNAs.
What we learned
A key lesson from this project is the importance of interpretability and understandability in designing computational tools for biology. Building a workflow that is both technically sound and transparent to users ensures broader accessibility and trust in the predictions generated.
What's next for CRISPRight
Looking ahead, we plan to improve model performance by incorporating more advanced heuristics and enforcing stricter biological constraints on guide RNA generation. This will enhance the robustness of the evolutionary simulations and move CRISPRight closer to a practical tool for safe and precise genome editing applications.


Log in or sign up for Devpost to join the conversation.