Inspiration
Our inspiration comes from wastewater surveillance for COVID-19. We were intrigued by how COVID-19 levels could be detected in a city or even a dorm building with no impact on daily life. Instead of major disruptions with test kits and nose swabs, data collection happens silently in the background. We set out to create a system that was more individual-focused and robust.
What it does
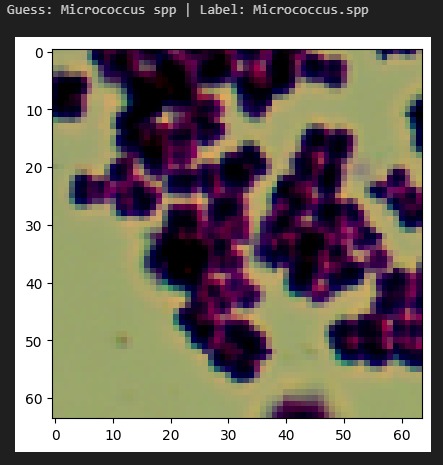
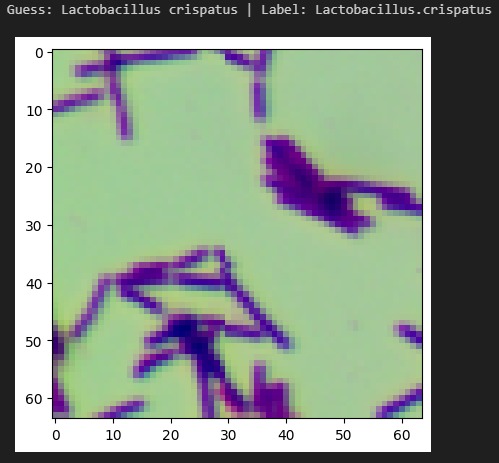
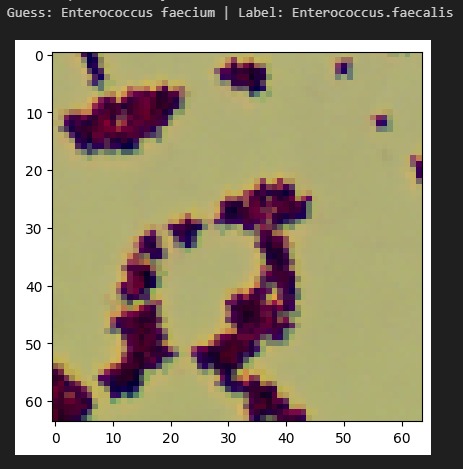
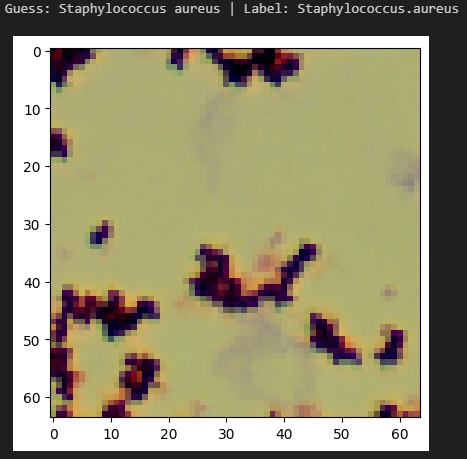
InfectaScan identifies bacteria based on microscope pictures, allowing healthcare professionals to continuously monitor patients' microbiome health and warn when antibiotics are being misused or the patient has acquired an opportunistic infection. Signs in the stool may result within 3 hours of colonization, a massive improvement over the days or weeks in which other symptoms would become apparent and a lab-based diagnosis made. This technology could lower the negative impact of prescribing the wrong antibiotics. With 44% of safety errors specifically attributed to medication errors like with antibiotics, especially in the elderly and in racial minorities, InfectaScan could massively improve patient safety, quality of life, and equity. It can currently distinguish 33 common species, including both beneficial species and species which are known to cause illness. We have the software components that will make a hardware component easy to implement and highly scalable.
How we built it
We used rapid prototyping techniques to build and optimize a CNN. This CNN was then containerized, which allowed us to run almost anywhere. We leveraged our team's experience with UIs to make an easy-to-use graphical interface.
Challenges we ran into
We had early challenges finding an appropriate dataset for our AI. Since imaging-based bacterial identification is a relatively recent advancement, only a handful of small datasets were applicable. We encountered several roadblocks when training our AI, but many of these provided important insights. For example, early versions of our model had difficulty distinguishing between different species of the same genus. We thought this was an architectural issue, but we realized that this is reflective of the real world. Similar bacteria look similar! We marched forward with greater data augmentation and higher resolution, creating a robust system.
Accomplishments that we're proud of
All of our team are relatively new to AI and applied computer science in general. We learned much of what we applied during the Hackathon. We were excited to see our CNN model perform with flying colors in testing, making the most of our limited training data.
What we learned
We learned the ins and outs of building a CNN, including the related topics of image processing and deploying ML models. We learned how to use GitHub to collaborate in real-time, sharing tools (Python notebooks) that were not only used to create our model but may help others understand how to improve it.
What's next for InfectaScan
InfectaScan has a lot of potential. Implemented with hardware like microcontrollers and microscope cameras, we see it as a cost-effective and fast tool for improving patient safety. We envision a hybrid cloud/hardware product that can address both the needs of healthcare institutions as well as at-risk populations. With a cloud-based service like Google Cloud, even institutions without heavy computing resources can have accurate diagnostic data. Additionally, we hope that lab-verified infection data can be fed back into our system, increasing the number of identifiable species.

Log in or sign up for Devpost to join the conversation.