This video provides examples of how you can extract and match patterns using regular expressions in R. This is a great tool for manipulating strings as needed.
Tag Archives: R programming

Fuzzy Joins with R
In this post, we will look at how you can make joins between datasets using a fuzzy criteria in which the matches between the datasets are not 100% the same. In order to do this, we will use the following packages in R.
library(stringdist)
library(stringr)
library(fuzzyjoin)The “stringdist” package will be used to measure the differences between various strings that we will use. The “stringr” package will be used to manipulate some text. Lastly, the “fuzzyjoin” package will be used to join datasets.
String Distance
The stringdist() function is used to measure the differences between strings. This is measured in many different ways. For our purposes, the distance is measured by the number of changes the function has to make so that the second string is the same as the original string of comparison. Below is code that uses three different methods each to compare the strings in the function.
> stringdist("darrin", "darren", method = "lv")
[1] 1
> stringdist("darrin", "darren", method = "dl")
[1] 1
> stringdist("darrin", "darren", method = "osa")
[1] 1This code is simple. First, we call the function. Inside the function, the first string is the ground truth string which is the string everything else is compared to. The second string is the other string that is compared to the first one. The method is how the difference is measured. For each example, we picked a different method. “lv” stands for Levenshtein distance, “dl” stands for full Damerau-Levenshtein distance, and “osa” stands for Optimal String Alignment distance. The details of each of these methods can be found by looking at the documentation for the “stringdist” package. Also, note that there are other methods beyond this that are available as well.
The value for these methods is 1, which means that only one change is needed to convert “darren” to “darrin”. Most of the time the methods are highly similar in their results but just to demonstrate, below is an example where the methods disagree.
> stringdist("darrin", "dorirn", method = "lv")
[1] 3
> stringdist("darrin", "dorirn", method = "dl")
[1] 2
> stringdist("darrin", "dorirn", method = "osa")
[1] 2Now, the values are different. The reason behind these differences is explained in the documentation.
amatch()
The amatch() function allows you to compare multiple strings to the ground truth and indicate which one is closest to the original ground truth string. Below is the code and output from this function.
amatch(
x = "Darrin",
table = c("Darren", "Daren", "Darien"),
maxDist = 1,
method = "lv"
)
[1] 1Here is what we did.
- The x argument is the ground truth. In other words, all other strings are compared to the value of x.
- The “table” argument contains all the strings that are being compared to the x argument.
- “maxDist” is how far away or how many changes max can be made in order for the strings in the “table” to be considered the best match
- “method” is the method used to calculate the “maxdist”
- The output is 1. This means that the first string in the table “Darren” has a max distance of 1
Fuzzy Join
The fuzzy join is used to join tables that have columns that are similar but not the same. Normally, joins work on exact matches but the fuzzy join does not require this. Before we use this function we have to prepare some data. We will modify the “Titanic” dataset to run this example. The “Titanic” dataset is a part of R by default and there is no need for any packages. Below is the code for the data preparation.
Titanic_1<-as.data.frame(Titanic)
Titanic_2<-as.data.frame(Titanic)
Titanic_1$Sex<-str_to_lower(Titanic_1$Sex)
Titanic_1$Age<-str_to_lower(Titanic_1$Age)Here is what we did.
- We saved two copies of the “Titanic” dataset as dataframes. This was done because the fuzzy join function needs dataframes.
- Next, we made clear differences between the two datasets. For “Titanic_1” we lowercase the sex, and age columns so that there was not an exact match when joining these two dataframes with the fuzzy join function.
We will now use the fuzzy join function. Below is the code followed by the output.
stringdist_join(
Titanic_1,
Titanic_2,
by = c("Age" = "Age","Sex"="Sex"),
method = "lv",
max_dist = 1,
distance_col = "distance"
)
The stringdist_join() function is used to perform the fuzzy join. “Titanic_1” is the x dataset and “Titanic_2” is the y dataset. The “by” argument tells the function which columns are being used in the join. The “method” argument indicates how the distance is calculated between the rows in each dataset. The “max_dist” argument is the criteria by which a join is made. In other words, if the distance is greater than one no join will take place. Lastly, the “distance_col” argument creates new columns that show the distance between the compared columns.
The output was a full join. All columns from both datasets are present. The columns with “.x” are from the “Titanic_1” while the columns with “.y” are from “Titanic_2”. The “.distance” column tells us the difference when that row of data was compared from each dataset. For example, in row 1 the “Age.distance” is 1. This means that the difference in “Age.x” and “Age.y” is 1. The only difference is that “Age.x” is lowercase while “Age.y” is capitalized.
Conclusion
The tools mentioned here allow you to match data that is different with a clear metric of the difference. This can be powerful when you have to join data that does not have a matching column in both datasets. Therefore, there is a place for the tools in the life of any data analyst who deals with fuzzy data like this.
Using glue_collapse() in R VIDEO

Glue Collapse in R
The glue_collapse() function is another powerful tool from the glue package. In this post, we will look at practical ways to use this function.
Collapsing Text
As you can probably tell from the name, the glue_collapse() function is useful for collapsing a string. In the code below, we will create an object containing several names and use glue_collapse to collapse the values into one string.
> library(glue)
> many_people<-c("Mike","Bob","James","Sam")
> glue_collapse(many_people)
MikeBobJamesSamIn the code above we called the glue package. Next, we created an object called “many_people” which contains several names separated by commas. Lastly, we called the glue_collapse() function which removes the quotes and commas from the string.
Separate & Last
Another great tool of the glue_collapse() function is the ability to separate strings and have a specific last argument. This technique helps to make the output highly readable. Below is the code
> glue_collapse(many_people,sep = ", ", last = ", and ")
Mike, Bob, James, and SamIn the code above we separate each string with a comma followed by a space in the “sep” argument. The “last” argument tells R what to do before the last word in the string. In this example, we have a comma followed by a space and the word and.
Collapse a Dataframe
The glue_collapse() function can also be used with data frames. In the example, below we will take a column from a dataframe and collapse it.
> head(iris$Species)
[1] setosa setosa setosa setosa setosa setosa
Levels: setosa versicolor virginica
> glue_collapse(iris$Species)
setosasetosasetosasetosasetosasetosaseto.....In the code above, we first take a look at the “Species” column from the “iris” dataset using the head() function. Next, we use the glue_collapse() function in the “Species” column. YOu can see how the rows are all collapsed into one long string in this example.
glue and glue_collapse working together
You can also use the glue() and glue_collapse function together as a team.
> glue(
+ "Hi {more_people}.",
+ more_people = glue_collapse(
+ many_people,sep = ", ",last = ", and "
+ )
+ )
Hi Mike, Bob, James, and Sam.This code is a little bit more complicated but here is what happened.
- On the outside, we start with the glue() function in the first line.
- Inside the glue() function we create a string that contains the word Hi and a temporary variable called “more_people”.
- Next, we define the temporary variable “more_people with the help of the glue_collapse() function.
- Inside the glue_collapse() function we separate the strings inside the “many_people” object.
As you can see, the use of the glue_collapse() and glue() functions can be endless.
Conclusion
The glue_collapse() function is another useful tool that can be used with text data. Knowing what options are available for data analysis makes the process much easier.
Using Glue in R VIDEO

Using Glue in R
The glue package in R provides a lot of great tools for using regular expressions and manipulating data. In this post, we will look at examples of using just the glue() function from this package.
Paste vs Glue
The paste() function is an older way of achieving the same things that we can achieve with the glue() function. paste() allows you to combine strings. Below we will load our packages and execute a command with the paste() function.
> library(glue)
> library(dplyr)
> people<-"Dan"
> paste("Hello",people)
[1] "Hello Dan"In the code above, we load the glue and the dplyr package (we will need dplyr later). We then create an object called “people” that contains the string “Dan”. We then used the past function to combine the “people” vector with the string “Hello”. The output is at the bottom of the code.
Below is an example of the same output but using the glue() function
> glue("Hello {people}")
Hello DanInside the glue() function everything is inside parentheses. However, the object “people” is inside curly braces and this indicates to the glue() function to look for what “people” represents. The printout is the same but without parentheses.
Multiple Strings
Below is an example of including multiple strings in the same glue() function
> people<-"Dan"
> people_2<-"Darrell"
> glue("Hello {people} and {people_2}")
Hello Dan and DarrellIn the first two lines above we make our objects. In line 3 we used the glue() function again and inside we included both objects in curly braces.
In another example using multiple strings we will replace text if it meets a certain criteria.
> people<-"Dan"
> people_2<-NA
> glue("Hi {people} and {people_2}",.na="What")
Hi Dan and WhatIn the code above we start by creating two objects. The second object (people_2) has stored NA. The code in the third line is the same with the exception of the “.na” argument. The “.na” argument is set to the string “What” which tells R to replace any NA values with the string “What”. The output is in the final line.
Temporary Variables
It is also possible to make variables that are temporary. The temporary variable can be named or unnamed. Below is an example with a named variable.
> glue("Dan is {height} cm tall.",height=175)
Dan is 175 cm tall.The temporary variable “height” is inside the curly braces. The value for “height” is set inside the function to 175.
It is also possible to have unnamed variables inside the function. Below we will use a function inside the curly braces.
> glue("The average number is {mean(c(2,3,4,5))}")
The average number is 3.5The example is self-explanatory. We used the mean() function inside the curly braces to get a calculated value. As you can see, the potential is endless.
Using Dataframes
In our last example, we will see how you can create a data frame and using input from one column to create a new column.
> df<-data.frame(column_1="Dan")
> df
column_1
1 Dan
> df %>% mutate(new_column = glue("Hi {column_1}"))
column_1 new_column
1 Dan Hi DanHere is what we did.
- We made a dataframe called df. This dataframe contains one column called column_1. In column_1 we have the string Dan.
- In line 2 we display the values of the dataframe.
- Next, we use the mutate() function to create a new column. Inside the mutate function we use the glue function and set it to create a string that uses the word “Hi” in front of the values of column_1.
- Lastly, we print out the results.
Conclusion
The glue package provides many powerful tools for manipulating data. The examples provided here only focus on one function. As such, this package is sure to provide useful ways in which data analyst can modify their data.
Intro to R Markdown VIDEO
R Markdown is a tool within RStudio that is beneficial for reporting results from an analysis that was done in RStudio. The video below provides several basics ways that R Markdown can be used in a document.


Linking Plots in Plotly with R ViDEO
Linking plots involves allowing the action you take in one plot to affect another. Doing this can allow the user to uncover various patterns that may not be apparent otherwise. Using plotly, it is possible to link plots and this is shown in the video below.


Animation with Plotly in R
Making Scatterplots Using Plotly in R
Making scatterplots is a part of the data analyst’s life. The video below shows how a scatterplot can be created using ploty in the R environment.


Making Bar Graphs with Plotly in R VIDEO
The video below provides examples of how to make bar graphs using plotly in R.


Histograms Using Plotly in R
Plotly is a library that allows you to make interactive charts in R. The example in the video below will focus on making histograms with this library.

Aggregating Data with dplyr VIDEO

More Selecting and Transforming with dplyR
In this post, we are going to learn some more advance ways to work with functions in the dplyr package. Let’s load our libraries
library(dplyr)
library(gapminder)Our dataset is the gapminder dataset which provides information about countries and continents related to gdp, life expectancy, and population. Here is what the data looks like as a refresher.
glimpse(gapminder)## Rows: 1,704
## Columns: 6
## $ country <fct> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", …
## $ continent <fct> Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, …
## $ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, …
## $ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.8…
## $ pop <int> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12…
## $ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, …select
You can use the colon symbol to select multiple columns at once. Doing this is a great way to save time when selecting variables.
gapminder%>%
select(lifeExp:gdpPercap)## # A tibble: 1,704 x 3
## lifeExp pop gdpPercap
## <dbl> <int> <dbl>
## 1 28.8 8425333 779.
## 2 30.3 9240934 821.
## 3 32.0 10267083 853.
## 4 34.0 11537966 836.
## 5 36.1 13079460 740.
## 6 38.4 14880372 786.
## 7 39.9 12881816 978.
## 8 40.8 13867957 852.
## 9 41.7 16317921 649.
## 10 41.8 22227415 635.
## # … with 1,694 more rowsYou can see that by using the colon we were able to select the last three columns.
There are also arguments called “select helpers.” Select helpers help you find columns in really large data sets. For example, let’s say we want columns that contain the string “life” in them. To find this we would use the contain argument as shown below.
gapminder%>%
select(contains('life'))## # A tibble: 1,704 x 1
## lifeExp
## <dbl>
## 1 28.8
## 2 30.3
## 3 32.0
## 4 34.0
## 5 36.1
## 6 38.4
## 7 39.9
## 8 40.8
## 9 41.7
## 10 41.8
## # … with 1,694 more rowsOnly the column that contains the string life is selected. There are other help selectors that you can try on your own such as starts_with, ends_with and more.
To remove a variable from a dataset you simply need to put a minus sign in front of it as shown below.
gapminder %>%
select(-lifeExp, -gdpPercap)## # A tibble: 1,704 x 4
## country continent year pop
## <fct> <fct> <int> <int>
## 1 Afghanistan Asia 1952 8425333
## 2 Afghanistan Asia 1957 9240934
## 3 Afghanistan Asia 1962 10267083
## 4 Afghanistan Asia 1967 11537966
## 5 Afghanistan Asia 1972 13079460
## 6 Afghanistan Asia 1977 14880372
## 7 Afghanistan Asia 1982 12881816
## 8 Afghanistan Asia 1987 13867957
## 9 Afghanistan Asia 1992 16317921
## 10 Afghanistan Asia 1997 22227415
## # … with 1,694 more rowsIn the output above you can see that life expectancy and per capa GDP are missing.
rename
Another function is the rename function which allows you to rename a variable. Below is an example in which the variable “pop” is renamed “population.”
gapminder %>%
select(country, year, pop) %>%
rename(population=pop)## # A tibble: 1,704 x 3
## country year population
## <fct> <int> <int>
## 1 Afghanistan 1952 8425333
## 2 Afghanistan 1957 9240934
## 3 Afghanistan 1962 10267083
## 4 Afghanistan 1967 11537966
## 5 Afghanistan 1972 13079460
## 6 Afghanistan 1977 14880372
## 7 Afghanistan 1982 12881816
## 8 Afghanistan 1987 13867957
## 9 Afghanistan 1992 16317921
## 10 Afghanistan 1997 22227415
## # … with 1,694 more rowsYou can see that the “pop” variable has been renamed. Remember that the new name goes on the left of the equal sign while the old name goes on the right of the equal sign.
There is a shortcut to this and it involves renaming variables inside the select function. In the example below, we rename the pop variable population inside the select function.
gapminder %>%
select(country, year, population=pop)## # A tibble: 1,704 x 3
## country year population
## <fct> <int> <int>
## 1 Afghanistan 1952 8425333
## 2 Afghanistan 1957 9240934
## 3 Afghanistan 1962 10267083
## 4 Afghanistan 1967 11537966
## 5 Afghanistan 1972 13079460
## 6 Afghanistan 1977 14880372
## 7 Afghanistan 1982 12881816
## 8 Afghanistan 1987 13867957
## 9 Afghanistan 1992 16317921
## 10 Afghanistan 1997 22227415
## # … with 1,694 more rowstransmute
The transmute function allows you to select and mutate variables at the same time. For example, let’s say that we want to know total gdp we could find this by multplying the population by gdp per capa. This is done with the transmute function below.
gapminder %>%
transmute(country, year, total_gdp = pop * gdpPercap)## # A tibble: 1,704 x 3
## country year total_gdp
## <fct> <int> <dbl>
## 1 Afghanistan 1952 6567086330.
## 2 Afghanistan 1957 7585448670.
## 3 Afghanistan 1962 8758855797.
## 4 Afghanistan 1967 9648014150.
## 5 Afghanistan 1972 9678553274.
## 6 Afghanistan 1977 11697659231.
## 7 Afghanistan 1982 12598563401.
## 8 Afghanistan 1987 11820990309.
## 9 Afghanistan 1992 10595901589.
## 10 Afghanistan 1997 14121995875.
## # … with 1,694 more rowsConclusion
With these basic tools it is now a little easier to do some data analysis when using R. There is so much more than can be learned but this will have to wait for the future.
Transform Variables with dplyR VIDEO

Data Aggregation with dplyr
In this post, we will learn about data aggregation with the dplyr package. Data aggregation is primarily a tool for summarizing the information you have collected. Let’s start by loading our packages.
library(dplyr)
library(gapminder)dplyr is for the data manipulation while gapminder provides us with the data. We will learn the following functions from the dplyr package
- count()
- summarize
- group_by()
- top_n()
count
The count function allows you to count the number of observations in your dataset as shown below.
gapminder %>%
count()## # A tibble: 1 x 1
## n
## <int>
## 1 1704The output tells us that there are over 1700 rows of data. However, the count function can do much more. For example, we can also count values in a specific column. Below, we calculated how many rows of data we have by continent.
gapminder %>%
count(continent)## # A tibble: 5 x 2
## continent n
## <fct> <int>
## 1 Africa 624
## 2 Americas 300
## 3 Asia 396
## 4 Europe 360
## 5 Oceania 24The output speaks for its self. There are two columns the left is continent and the right is how many times that particular continent appears in the dataset. You can also sort this data by adding the argument called sort as shown below.
gapminder %>%
count(continent, sort =TRUE)## # A tibble: 5 x 2
## continent n
## <fct> <int>
## 1 Africa 624
## 2 Asia 396
## 3 Europe 360
## 4 Americas 300
## 5 Oceania 24There is another argument we can add and this is called the weight or wt argument. The wt argument adds up the values of the population in our example and we can now see how many respondents there were from each continent. Below is the code an example
gapminder %>%
count(continent, wt=pop, sort=TRUE)## # A tibble: 5 x 2
## continent n
## <fct> <dbl>
## 1 Asia 30507333901
## 2 Americas 7351438499
## 3 Africa 6187585961
## 4 Europe 6181115304
## 5 Oceania 212992136You can see that we now know how many people from each continent were in the dataset.
summarize
The summarize function takes many rows of data and reduce it to a single output. For example, if we want to know the total number of people in the dataset we could run the code below.
gapminder %>%
summarize(total_pop=sum(pop))## # A tibble: 1 x 1
## total_pop
## <dbl>
## 1 50440465801You can also continue to add more and more things you want to know be separating them with a comma. In the code below, we add to it the average GDP.
gapminder %>%
summarize(total_pop=sum(pop), average_gdp=mean(gdpPercap))## # A tibble: 1 x 2
## total_pop average_gdp
## <dbl> <dbl>
## 1 50440465801 7215.group_by
The group by function allows you to aggregate data by groups. For example, if we want to know the total population and the average gdp by continent the code below would help to learn this.
gapminder %>%
group_by(continent) %>%
summarize(total_pop=sum(pop), mean_gdp=mean(gdpPercap)) %>%
arrange(desc(total_pop))## # A tibble: 5 x 3
## continent total_pop mean_gdp
## <fct> <dbl> <dbl>
## 1 Asia 30507333901 7902.
## 2 Americas 7351438499 7136.
## 3 Africa 6187585961 2194.
## 4 Europe 6181115304 14469.
## 5 Oceania 212992136 18622.It is also possible to group by more than one column. However, to do this we need to create another categorical variable. We are going to use mutate to create a categorical variable that breaks the data into two parts. Before 1980 and after 1980. Then we will group by country and whether the mean of the gdp was collected before or after 1980. Below is the code
gapminder %>%
mutate(before_1980=if_else(year < 1980, "yes","no")) %>%
group_by(country, before_1980) %>%
summarize(mean_gdp=mean(gdpPercap))## # A tibble: 284 x 3
## # Groups: country [142]
## country before_1980 mean_gdp
## <fct> <chr> <dbl>
## 1 Afghanistan no 803.
## 2 Afghanistan yes 803.
## 3 Albania no 3934.
## 4 Albania yes 2577.
## 5 Algeria no 5460.
## 6 Algeria yes 3392.
## 7 Angola no 2944.
## 8 Angola yes 4270.
## 9 Argentina no 9998.
## 10 Argentina yes 7913.
## # … with 274 more rowstop_n
The top_n function allows you to find the most extreme values when looking at groups. For example, we could find which countries has the highest life expectancy by continent. The answer is below
gapminder %>%
group_by(continent) %>%
top_n(1, lifeExp)## # A tibble: 5 x 6
## # Groups: continent [5]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Australia Oceania 2007 81.2 20434176 34435.
## 2 Canada Americas 2007 80.7 33390141 36319.
## 3 Iceland Europe 2007 81.8 301931 36181.
## 4 Japan Asia 2007 82.6 127467972 31656.
## 5 Reunion Africa 2007 76.4 798094 7670.As an example, Japan has the highest life expectancy in Asia. Canada has the highest life expectancy in the Americas. Naturally you are not limited to the top 1. This number can be changed to whatever you want. For example, below we change the number to 3.
gapminder %>%
group_by(continent) %>%
top_n(3, lifeExp)## # A tibble: 15 x 6
## # Groups: continent [5]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Australia Oceania 2002 80.4 19546792 30688.
## 2 Australia Oceania 2007 81.2 20434176 34435.
## 3 Canada Americas 2002 79.8 31902268 33329.
## 4 Canada Americas 2007 80.7 33390141 36319.
## 5 Costa Rica Americas 2007 78.8 4133884 9645.
## 6 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 7 Iceland Europe 2007 81.8 301931 36181.
## 8 Japan Asia 2002 82 127065841 28605.
## 9 Japan Asia 2007 82.6 127467972 31656.
## 10 New Zealand Oceania 2007 80.2 4115771 25185.
## 11 Reunion Africa 1997 74.8 684810 6072.
## 12 Reunion Africa 2002 75.7 743981 6316.
## 13 Reunion Africa 2007 76.4 798094 7670.
## 14 Spain Europe 2007 80.9 40448191 28821.
## 15 Switzerland Europe 2007 81.7 7554661 37506.
Transform Data with dplyr
In this post, we will be exposed to tools for wrangling and manipulating data in R.
Let’s begin by loading the libraries we will be using. We will use the dplyr package and the gapminder package. dplyr is for manipulating the data and gapminder provides the dataset.
library(dplyr)
library(gapminder)You can look at the data briefly by using a function called “glimpse” as shown below.
glimpse(gapminder)## Rows: 1,704
## Columns: 6
## $ country <fct> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", …
## $ continent <fct> Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, …
## $ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, …
## $ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.8…
## $ pop <int> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12…
## $ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, …You can see that we have six columns or variables and over 1700 rows of data. This data provides information about countries and various demographic statistics.
select()
The select function allows you to grab only the variables you want for analysis. This becomes exceptionally important when you have a large number of variables. In our next example, we will select 4 variables from the gapminder dataset. Below is the code to achieve this.
gapminder %>%
select(country,continent, pop, lifeExp)## # A tibble: 1,704 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 Afghanistan Asia 8425333 28.8
## 2 Afghanistan Asia 9240934 30.3
## 3 Afghanistan Asia 10267083 32.0
## 4 Afghanistan Asia 11537966 34.0
## 5 Afghanistan Asia 13079460 36.1
## 6 Afghanistan Asia 14880372 38.4
## 7 Afghanistan Asia 12881816 39.9
## 8 Afghanistan Asia 13867957 40.8
## 9 Afghanistan Asia 16317921 41.7
## 10 Afghanistan Asia 22227415 41.8
## # … with 1,694 more rowsThe strange symbol %>% is called a “pipe” and allows you to continuously build your code. You can also save this information by assigning a name to an object like any other variable in r.
country_data<-gapminder %>%
select(country,continent, pop, lifeExp)arrange
The arrange verb sorts your data based on one or more variables. For example, let’s say we want to know which country has the highest population. The code below provides the answer.
country_data %>%
arrange(pop)## # A tibble: 1,704 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 Sao Tome and Principe Africa 60011 46.5
## 2 Sao Tome and Principe Africa 61325 48.9
## 3 Djibouti Africa 63149 34.8
## 4 Sao Tome and Principe Africa 65345 51.9
## 5 Sao Tome and Principe Africa 70787 54.4
## 6 Djibouti Africa 71851 37.3
## 7 Sao Tome and Principe Africa 76595 56.5
## 8 Sao Tome and Principe Africa 86796 58.6
## 9 Djibouti Africa 89898 39.7
## 10 Sao Tome and Principe Africa 98593 60.4
## # … with 1,694 more rowsTo complete this task we had to use the arrange function and place the name of the variable we want to sort by inside the parentheses. However, this is not exactly what we want. What we have found is the countries with the smallest population. To sort from largest to smallest you must use the desc function as well and this is shown below.
country_data %>%
arrange(desc(pop))## # A tibble: 1,704 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 China Asia 1318683096 73.0
## 2 China Asia 1280400000 72.0
## 3 China Asia 1230075000 70.4
## 4 China Asia 1164970000 68.7
## 5 India Asia 1110396331 64.7
## 6 China Asia 1084035000 67.3
## 7 India Asia 1034172547 62.9
## 8 China Asia 1000281000 65.5
## 9 India Asia 959000000 61.8
## 10 China Asia 943455000 64.0
## # … with 1,694 more rowsNow, this is what we want. China claims several of the top spots. The reason a country is on the list more than once is that the data was collected several different years.
filter
The filter function is used to obtain only specific values that meet the criteria. For example, what if we want to know the population of only India in descending order. Below is the code for how to do this.
country_data %>%
arrange(desc(pop)) %>%
filter(country=='India') ## # A tibble: 12 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 India Asia 1110396331 64.7
## 2 India Asia 1034172547 62.9
## 3 India Asia 959000000 61.8
## 4 India Asia 872000000 60.2
## 5 India Asia 788000000 58.6
## 6 India Asia 708000000 56.6
## 7 India Asia 634000000 54.2
## 8 India Asia 567000000 50.7
## 9 India Asia 506000000 47.2
## 10 India Asia 454000000 43.6
## 11 India Asia 409000000 40.2
## 12 India Asia 372000000 37.4Now we have only data that relates to India. All we did was include one more pipe and the filter function. We had to tell R which country by placing the information above in the parentheses.
filter is not limited to text searches. You can also search based on numerical values. For example, what if we only want countries with a life expectancy of 81 or higher
country_data %>%
arrange(desc(pop)) %>%
filter(lifeExp >= 81)## # A tibble: 7 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 Japan Asia 127467972 82.6
## 2 Japan Asia 127065841 82
## 3 Australia Oceania 20434176 81.2
## 4 Switzerland Europe 7554661 81.7
## 5 Hong Kong, China Asia 6980412 82.2
## 6 Hong Kong, China Asia 6762476 81.5
## 7 Iceland Europe 301931 81.8You can see the results for yourself. It is also possible to combine multiply conditions for whatever functions are involved. For example, if I want to arrange my data by population and country while also filtering it by a population greater than 100,000,000,000 and with a life expectancy of less than 45. This is shown below
country_data %>%
arrange(desc(pop, country)) %>%
filter(pop>100000000, lifeExp<45)## # A tibble: 5 x 4
## country continent pop lifeExp
## <fct> <fct> <int> <dbl>
## 1 China Asia 665770000 44.5
## 2 China Asia 556263527 44
## 3 India Asia 454000000 43.6
## 4 India Asia 409000000 40.2
## 5 India Asia 372000000 37.4mutate
The mutate function is for manipulating variables and creating new ones. For example, the gdpPercap variable is highly skewed. We can create a variable of gdpercap that is the log of this variable. Using the log will help the data to assume the characteristics of a normal distribution. Below is the code for this.
gapminder %>%
select(country,continent, pop, gdpPercap) %>%
mutate(log_gdp=log(gdpPercap))## # A tibble: 1,704 x 5
## country continent pop gdpPercap log_gdp
## <fct> <fct> <int> <dbl> <dbl>
## 1 Afghanistan Asia 8425333 779. 6.66
## 2 Afghanistan Asia 9240934 821. 6.71
## 3 Afghanistan Asia 10267083 853. 6.75
## 4 Afghanistan Asia 11537966 836. 6.73
## 5 Afghanistan Asia 13079460 740. 6.61
## 6 Afghanistan Asia 14880372 786. 6.67
## 7 Afghanistan Asia 12881816 978. 6.89
## 8 Afghanistan Asia 13867957 852. 6.75
## 9 Afghanistan Asia 16317921 649. 6.48
## 10 Afghanistan Asia 22227415 635. 6.45
## # … with 1,694 more rowsIn the code above we had to select our variables again and then we create the new variable “log_gdp”. This new variable appears all the way to the right in the dataset. Naturally, we can extend our code by using our new variable in other functions as shown below.
Conclusion
This post was longer than normal but several practical things were learned. You now know some basic techniques for wrangling data using the dplyr package in R.

T-SNE Visualization and R
It is common in research to want to visualize data in order to search for patterns. When the number of features increases, this can often become even more important. Common tools for visualizing numerous features include principal component analysis and linear discriminant analysis. Not only do these tools work for visualization they can also be beneficial in dimension reduction.
However, the available tools for us are not limited to these two options. Another option for achieving either of these goals is t-Distributed Stochastic Embedding. This relative young algorithm (2008) is the focus of the post. We will explain what it is and provide an example using a simple dataset from the Ecdat package in R.
t-sne Defined
t-sne is a nonlinear dimension reduction visualization tool. Essentially what it does is identify observed clusters. However, it is not a clustering algorithm because it reduces the dimensions (normally to 2) for visualizing. This means that the input features are not longer present in their original form and this limits the ability to make inference. Therefore, t-sne is often used for exploratory purposes.
T-sne non-linear characteristic is what makes it often appear to be superior to PCA, which is linear. Without getting too technical t-sne takes simultaneously a global and local approach to mapping points while PCA can only use a global approach.
The downside to t-sne approach is that it requires a large amount of calculations. The calculations are often pairwise comparisons which can grow exponential in large datasets.
Initial Packages
We will use the “Rtsne” package for the analysis, and we will use the “Fair” dataset from the “Ecdat” package. The “Fair” dataset is data collected from people who had cheated on their spouse. We want to see if we can find patterns among the unfaithful people based on their occupation. Below is some initial code.
library(Rtsne)
library(Ecdat)Dataset Preparation
To prepare the data, we first remove in rows with missing data using the “na.omit” function. This is saved in a new object called “train”. Next, we change or outcome variable into a factor variable. The categories range from 1 to 9
- Farm laborer, day laborer,
- Unskilled worker, service worker,
- Machine operator, semiskilled worker,
- Skilled manual worker, craftsman, police,
- Clerical/sales, small farm owner,
- Technician, semiprofessional, supervisor,
- Small business owner, farm owner, teacher,
- Mid-level manager or professional,
- Senior manager or professional.
Below is the code.
train<-na.omit(Fair)
train$occupation<-as.factor(train$occupation)Visualization Preparation
Before we do the analysis we need to set the colors for the different categories. This is done with the code below.
colors<-rainbow(length(unique(train$occupation)))
names(colors)<-unique(train$occupation)We can now do are analysis. We will use the “Rtsne” function. When you input the dataset you must exclude the dependent variable as well as any other factor variables. You also set the dimensions and the perplexity. Perplexity determines how many neighbors are used to determine the location of the datapoint after the calculations. Verbose just provides information during the calculation. This is useful if you want to know what progress is being made. max_iter is the number of iterations to take to complete the analysis and check_duplicates checks for duplicates which could be a problem in the analysis. Below is the code.
tsne<-Rtsne(train[,-c(1,4,7)],dims=2,perplexity=30,verbose=T,max_iter=1500,check_duplicates=F)## Performing PCA
## Read the 601 x 6 data matrix successfully!
## OpenMP is working. 1 threads.
## Using no_dims = 2, perplexity = 30.000000, and theta = 0.500000
## Computing input similarities...
## Building tree...
## Done in 0.05 seconds (sparsity = 0.190597)!
## Learning embedding...
## Iteration 1450: error is 0.280471 (50 iterations in 0.07 seconds)
## Iteration 1500: error is 0.279962 (50 iterations in 0.07 seconds)
## Fitting performed in 2.21 seconds.Below is the code for making the visual.
plot(tsne$Y,t='n',main='tsne',xlim=c(-30,30),ylim=c(-30,30))
text(tsne$Y,labels=train$occupation,col = colors[train$occupation])
legend(25,5,legend=unique(train$occupation),col = colors,,pch=c(1))
You can see that there are clusters however, the clusters are all mixed with the different occupations. What this indicates is that the features we used to make the two dimensions do not discriminant between the different occupations.
Conclusion
T-SNE is an improved way to visualize data. This is not to say that there is no place for PCA anymore. Rather, this newer approach provides a different way of quickly visualizing complex data without the limitations of PCA.

Web Scraping with R
In this post we are going to learn how to do web scrapping with R.Web scraping is a process for extracting data from a website. We have all done web scraping before. For example, whenever you copy and paste something from a website into another document such as Word this is an example of web scraping. Technically, this is an example of manual web scraping. The main problem with manual web scraping is that it is labor intensive and takes a great deal of time.
Another problem with web scraping is that the data can come in an unstructured manner. This means that you have to organize it in some way in order to conduct a meaningful analysis. This also means that you must have a clear purpose for what you are scraping along with answerable questions. Otherwise, it is easy to become confused quickly when web scraping
Therefore, we will learn how to automate this process using R. We will need the help of the “rest” and “xml2” packages to do this. Below is some initial code
library(rvest);library(xml2)For our example, we are going to scrape the titles and prices of books from a webpage on Amazon. When simply want to make an organized data frame. The first thing we need to do is load the URL into R and have R read the website using the “read_html” function. The code is below.
url<-'https://www.amazon.com/s/ref=nb_sb_noss?url=search-alias%3Daps&field-keywords=books'
webpage<-read_html(url)We now need to specifically harvest the titles from the webpage as this is one of our goals. There are at least two ways to do this. If you are an expert in HTML you can find the information by inspecting the page’s HTML. Another way is to the selectorGadget extension available in Chrome. When using this extension you simply click on the information you want to inspect and it gives you the CSS selector for that particular element. This is shown below
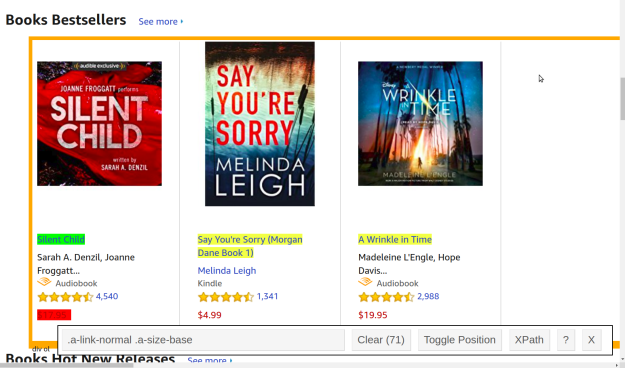
The green highlight is the CSS selector that you clicked on. The yellow represents all other elements that have the same CSS selector. The red represents what you do not want to be included. In this picture, I do not want the price because I want to scrape this separately.
Once you find your information you want to copy the CSS element information in the bar at the bottom of the picture. This information is then pasted into R and use the “html_nodes” function to pull this specific information from the webpage.
bookTitle<- html_nodes(webpage,'.a-link-normal .a-size-base')We now need to convert this information to text and we are done.
title <- html_text(bookTitle, trim = TRUE) Next, we repeat this process for the price.
bookPrice<- html_nodes(webpage,'.acs_product-price__buying')
price <- html_text(bookPrice, trim = TRUE) Lastly, we make our data frame with all of our information.
books<-as.data.frame(title)
books$price<-priceWith this done we can do basic statistical analysis such as the mean, standard deviation, histogram, etc. This was not a complex example but the basics of pulling data was provided. Below is what the first few entries of the data frame look like.
head(books)
## title price
## 1 Silent Child $17.95
## 2 Say You're Sorry (Morgan Dane Book 1) $4.99
## 3 A Wrinkle in Time $19.95
## 4 The Whiskey Sea $3.99
## 5 Speaking in Bones: A Novel $2.99
## 6 Harry Potter and the Sorcerer's Stone $8.99Conclusion
Web scraping using automated tools saves time and increases the possibilities of data analysis. The most important thing to remember is to understand what exactly it is you want to know. Otherwise, you will quickly become lost due to the overwhelming amounts of available information.
aggregate Function in R VIDEO
Using the aggregate function in R.
Creating Subgroups of Data in R VIDEO
Create subgroups in R
Subsetting Data in R VIDEO
Subsetting data in R
Getting Data Out of R Video
Getting data out of R
Importing Data into R VIDEO
Importing data into R
for loops in R VIDEO
for loops in R
APA Tables in R
Anybody who has ever had to do any writing for academic purposes or in industry has had to deal with APA formatting. The rules and expectations seem to be endless and always changing. If you are able to maneuver the endless list of rules you still have to determine what to report and how when writing an article.
There is a package in R that can at least take away the mystery of how to report ANOVA, correlation, and regression tables. This package is called “apaTables”. In this post, we will look at how to use this package for making tables that are formatted according to APA.
We are going to create examples of ANOVA, correlation, and regression tables using the ‘mtcars’ dataset. Below is the initial code that we need to begin.
library(apaTables)
data("mtcars")ANOVA
We will begin with the results of ANOVA. In order for this to be successful, you have to use the “lm” function to create the model. If you are familiar with ANOVA and regression this should not be surprising as they both find the same answer using different approaches. After the “lm” function you must use the “filename” argument and give the output a name in quotations. This file will be saved in your R working directory. You can also provide other information such as the table number and confidence level if you desire.
There will be two outputs in our code. The output to the console is in R. A second output will be in a word doc. Below is the code.
apa.aov.table(lm(mpg~cyl,mtcars),filename = "Example1.doc",table.number = 1)##
##
## Table 1
##
## ANOVA results using mpg as the dependent variable
##
##
## Predictor SS df MS F p partial_eta2
## (Intercept) 3429.84 1 3429.84 333.71 .000
## cyl 817.71 1 817.71 79.56 .000 .73
## Error 308.33 30 10.28
## CI_90_partial_eta2
##
## [.56, .80]
##
##
## Note: Values in square brackets indicate the bounds of the 90% confidence interval for partial eta-squaredHere is the word doc output

Perhaps you are beginning to see the beauty of using this package and its functions. The “apa.aov.table”” function provides a nice table that requires no formatting by the researcher.
You can even make a table of the means and standard deviations of ANOVA. This is similar to what you would get if you used the “aggregate” function. Below is the code.
apa.1way.table(cyl, mpg,mtcars,filename = "Example2.doc",table.number = 2)##
##
## Table 2
##
## Descriptive statistics for mpg as a function of cyl.
##
## cyl M SD
## 4 26.66 4.51
## 6 19.74 1.45
## 8 15.10 2.56
##
## Note. M and SD represent mean and standard deviation, respectively.
## Here is what it looks like in word

Correlation
We will now look at an example of a correlation table. The function for this is “apa.cor.table”. This function works best with only a few variables. Otherwise, the table becomes bigger than a single sheet of paper. In addition, you probably will want to suppress the confidence intervals to save space. There are other arguments that you can explore on your own. Below is the code
apa.cor.table(mtcars,filename = "Example3.doc",table.number = 3,show.conf.interval = F)##
##
## Table 3
##
## Means, standard deviations, and correlations
##
##
## Variable M SD 1 2 3 4 5 6 7
## 1. mpg 20.09 6.03
##
## 2. cyl 6.19 1.79 -.85**
##
## 3. disp 230.72 123.94 -.85** .90**
##
## 4. hp 146.69 68.56 -.78** .83** .79**
##
## 5. drat 3.60 0.53 .68** -.70** -.71** -.45**
##
## 6. wt 3.22 0.98 -.87** .78** .89** .66** -.71**
##
## 7. qsec 17.85 1.79 .42* -.59** -.43* -.71** .09 -.17
##
## 8. vs 0.44 0.50 .66** -.81** -.71** -.72** .44* -.55** .74**
##
## 9. am 0.41 0.50 .60** -.52** -.59** -.24 .71** -.69** -.23
##
## 10. gear 3.69 0.74 .48** -.49** -.56** -.13 .70** -.58** -.21
##
## 11. carb 2.81 1.62 -.55** .53** .39* .75** -.09 .43* -.66**
##
## 8 9 10
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## .17
##
## .21 .79**
##
## -.57** .06 .27
##
##
## Note. * indicates p < .05; ** indicates p < .01.
## M and SD are used to represent mean and standard deviation, respectively.
## Here is the word doc results

If you run this code at home and open the word doc in Word you will not see variables 9 and 10 because the table is too big by itself for a single page. I hade to resize it manually. One way to get around this is to delate the M and SD column and place those as rows below the table.
Regression
Our final example will be a regression table. The code is as follows
apa.reg.table(lm(mpg~disp,mtcars),filename = "Example4",table.number = 4)##
##
## Table 4
##
## Regression results using mpg as the criterion
##
##
## Predictor b b_95%_CI beta beta_95%_CI sr2 sr2_95%_CI
## (Intercept) 29.60** [27.09, 32.11]
## disp -0.04** [-0.05, -0.03] -0.85 [-1.05, -0.65] .72 [.51, .81]
##
##
##
## r Fit
##
## -.85**
## R2 = .718**
## 95% CI[.51,.81]
##
##
## Note. * indicates p < .05; ** indicates p < .01.
## A significant b-weight indicates the beta-weight and semi-partial correlation are also significant.
## b represents unstandardized regression weights; beta indicates the standardized regression weights;
## sr2 represents the semi-partial correlation squared; r represents the zero-order correlation.
## Square brackets are used to enclose the lower and upper limits of a confidence interval.
## Here are the results in word

You can also make regression tables that have multiple blocks or models. Below is an example
apa.reg.table(lm(mpg~disp,mtcars),lm(mpg~disp+hp,mtcars),filename = "Example5",table.number = 5)##
##
## Table 5
##
## Regression results using mpg as the criterion
##
##
## Predictor b b_95%_CI beta beta_95%_CI sr2 sr2_95%_CI
## (Intercept) 29.60** [27.09, 32.11]
## disp -0.04** [-0.05, -0.03] -0.85 [-1.05, -0.65] .72 [.51, .81]
##
##
##
## (Intercept) 30.74** [28.01, 33.46]
## disp -0.03** [-0.05, -0.02] -0.62 [-0.94, -0.31] .15 [.00, .29]
## hp -0.02 [-0.05, 0.00] -0.28 [-0.59, 0.03] .03 [-.03, .09]
##
##
##
## r Fit Difference
##
## -.85**
## R2 = .718**
## 95% CI[.51,.81]
##
##
## -.85**
## -.78**
## R2 = .748** Delta R2 = .03
## 95% CI[.54,.83] 95% CI[-.03, .09]
##
##
## Note. * indicates p < .05; ** indicates p < .01.
## A significant b-weight indicates the beta-weight and semi-partial correlation are also significant.
## b represents unstandardized regression weights; beta indicates the standardized regression weights;
## sr2 represents the semi-partial correlation squared; r represents the zero-order correlation.
## Square brackets are used to enclose the lower and upper limits of a confidence interval.
## Here is the word doc version

Conculsion
This is a real time saver for those of us who need to write and share statistical information.
Vectorization of function in R VIDEO
Vectorization of function in R
If Else Statements in Functions R VIDEO
If else statements in functions in R
If Statements with Functions in R VIDEO
If Statements in R
Arguments and Functions in R VIDEO
Arguments and functions in R
Intro to Functions in R VIDEOS
R functions intro
Intro to List VIDEO
Introduction to using List
Manipulating Dataframes in R VIDEO
Manipulating Dataframes in R
Intro to Dataframes in R VIDEO
Intro to dataframes in R
Local Regression in R
Local regression uses something similar to nearest neighbor classification to generate a regression line. In local regression, nearby observations are used to fit the line rather than all observations. It is necessary to indicate the percentage of the observations you want R to use for fitting the local line. The name for this hyperparameter is the span. The higher the span the smoother the line becomes.
Local regression is great one there are only a handful of independent variables in the model. When the total number of variables becomes too numerous the model will struggle. As such, we will only fit a bivariate model. This will allow us to process the model and to visualize it.
In this post, we will use the “Clothing” dataset from the “Ecdat” package and we will examine innovation (inv2) relationship with total sales (tsales). Below is some initial code.
library(Ecdat)data(Clothing)
str(Clothing)## 'data.frame': 400 obs. of 13 variables:
## $ tsales : int 750000 1926395 1250000 694227 750000 400000 1300000 495340 1200000 495340 ...
## $ sales : num 4412 4281 4167 2670 15000 ...
## $ margin : num 41 39 40 40 44 41 39 28 41 37 ...
## $ nown : num 1 2 1 1 2 ...
## $ nfull : num 1 2 2 1 1.96 ...
## $ npart : num 1 3 2.22 1.28 1.28 ...
## $ naux : num 1.54 1.54 1.41 1.37 1.37 ...
## $ hoursw : int 76 192 114 100 104 72 161 80 158 87 ...
## $ hourspw: num 16.8 22.5 17.2 21.5 15.7 ...
## $ inv1 : num 17167 17167 292857 22207 22207 ...
## $ inv2 : num 27177 27177 71571 15000 10000 ...
## $ ssize : int 170 450 300 260 50 90 400 100 450 75 ...
## $ start : num 41 39 40 40 44 41 39 28 41 37 ...There is no data preparation in this example. The first thing we will do is fit two different models that have different values for the span hyperparameter. “fit” will have a span of .41 which means it will use 41% of the nearest examples. “fit2” will use .82. Below is the code.
fit<-loess(tsales~inv2,span = .41,data = Clothing)
fit2<-loess(tsales~inv2,span = .82,data = Clothing)In the code above, we used the “loess” function to fit the model. The “span” argument was set to .41 and .82.
We now need to prepare for the visualization. We begin by using the “range” function to find the distance from the lowest to the highest value. Then use the “seq” function to create a grid. Below is the code.
inv2lims<-range(Clothing$inv2)
inv2.grid<-seq(from=inv2lims[1],to=inv2lims[2])The information in the code above is for setting our x-axis in the plot. We are now ready to fit our model. We will fit the models and draw each regression line.
plot(Clothing$inv2,Clothing$tsales,xlim=inv2lims)
lines(inv2.grid,predict(fit,data.frame(inv2=inv2.grid)),col='blue',lwd=3)
lines(inv2.grid,predict(fit2,data.frame(inv2=inv2.grid)),col='red',lwd=3)
Not much difference in the two models. For our final task, we will predict with our “fit” model using all possible values of “inv2” and also fit the confidence interval lines.
pred<-predict(fit,newdata=inv2.grid,se=T)
plot(Clothing$inv2,Clothing$tsales)
lines(inv2.grid,pred$fit,col='red',lwd=3)
lines(inv2.grid,pred$fit+2*pred$se.fit,lty="dashed",lwd=2,col='blue')
lines(inv2.grid,pred$fit-2*pred$se.fit,lty="dashed",lwd=2,col='blue')
Conclusion
Local regression provides another way to model complex non-linear relationships in low dimensions. The example here provides just the basics of how this is done is much more complicated than described here.
Smoothing Splines in R
This post will provide information on smoothing splines. Smoothing splines are used in regression when we want to reduce the residual sum of squares by adding more flexibility to the regression line without allowing too much overfitting.
In order to do this, we must tune the parameter called the smoothing spline. The smoothing spline is essentially a natural cubic spline with a knot at every unique value of x in the model. Having this many knots can lead to severe overfitting. This is corrected for by controlling the degrees of freedom through the parameter called lambda. You can manually set this value or select it through cross-validation.
We will now look at an example of the use of smoothing splines with the “Clothing” dataset from the “Ecdat” package. We want to predict “tsales” based on the use of innovation in the stores. Below is some initial code.
library(Ecdat)data(Clothing)
str(Clothing)## 'data.frame': 400 obs. of 13 variables:
## $ tsales : int 750000 1926395 1250000 694227 750000 400000 1300000 495340 1200000 495340 ...
## $ sales : num 4412 4281 4167 2670 15000 ...
## $ margin : num 41 39 40 40 44 41 39 28 41 37 ...
## $ nown : num 1 2 1 1 2 ...
## $ nfull : num 1 2 2 1 1.96 ...
## $ npart : num 1 3 2.22 1.28 1.28 ...
## $ naux : num 1.54 1.54 1.41 1.37 1.37 ...
## $ hoursw : int 76 192 114 100 104 72 161 80 158 87 ...
## $ hourspw: num 16.8 22.5 17.2 21.5 15.7 ...
## $ inv1 : num 17167 17167 292857 22207 22207 ...
## $ inv2 : num 27177 27177 71571 15000 10000 ...
## $ ssize : int 170 450 300 260 50 90 400 100 450 75 ...
## $ start : num 41 39 40 40 44 41 39 28 41 37 ...We are going to create three models. Model one will have 70 degrees of freedom, model two will have 7, and model three will have the number of degrees of freedom are determined through cross-validation. Below is the code.
fit1<-smooth.spline(Clothing$inv2,Clothing$tsales,df=57)
fit2<-smooth.spline(Clothing$inv2,Clothing$tsales,df=7)
fit3<-smooth.spline(Clothing$inv2,Clothing$tsales,cv=T)## Warning in smooth.spline(Clothing$inv2, Clothing$tsales, cv = T): cross-
## validation with non-unique 'x' values seems doubtful(data.frame(fit1$df,fit2$df,fit3$df))## fit1.df fit2.df fit3.df
## 1 57 7.000957 2.791762In the code above we used the “smooth.spline” function which comes with base r.Notice that we did not use the same coding syntax as the “lm” function calls for. The code above also indicates the degrees of freedom for each model. You can see that for “fit3” the cross-validation determine that 2.79 was the most appropriate degrees of freedom. In addition, if you type in the following code.
sapply(data.frame(fit1$x,fit2$x,fit3$x),length)## fit1.x fit2.x fit3.x
## 73 73 73You will see that there are only 73 data points in each model. The “Clothing” dataset has 400 examples in it. The reason for this reduction is that the “smooth.spline” function only takes unique values from the original dataset. As such, though there are 400 examples in the dataset only 73 of them are unique.
Next, we plot our data and add regression lines
plot(Clothing$inv2,Clothing$tsales)
lines(fit1,col='red',lwd=3)
lines(fit2,col='green',lwd=3)
lines(fit3,col='blue',lwd=3)
legend('topright',lty=1,col=c('red','green','blue'),c("df = 57",'df=7','df=CV 2.8'))
You can see that as the degrees of freedom increase so does the flexibility in the line. The advantage of smoothing splines is to have a more flexible way to assess the characteristics of a dataset.
Polynomial Spline Regression in R
Normally, when least squares regression is used, you fit one line to the model. However, sometimes you may want enough flexibility that you fit different lines over different regions of your independent variable. This process of fitting different lines over different regions of X is known as Regression Splines.
How this works is that there are different coefficient values based on the regions of X. As the researcher, you can set the cutoff points for each region. The cutoff point is called a “knot.” The more knots you use the more flexible the model becomes because there are fewer data points with each range allowing for more variability.
We will now go through an example of polynomial regression splines. Remeber that polynomial means that we will have a curved line as we are using higher order polynomials. Our goal will be to predict total sales based on the amount of innovation a store employs. We will use the “Ecdat” package and the “Clothing” dataset. In addition, we will need the “splines” package. The code is as follows.
library(splines);library(Ecdat)data(Clothing)We will now fit our model. We must indicate the number and placement of the knots. This is commonly down at the 25th 50th and 75th percentile. Below is the code
fit<-lm(tsales~bs(inv2,knots = c(12000,60000,150000)),data = Clothing)In the code above we used the traditional “lm” function to set the model. However, we also used the “bs” function which allows us to create our spline regression model. The argument “knots” was set to have three different values. Lastly, the dataset was indicated.
Remember that the default spline model in R is a third-degree polynomial. This is because it is hard for the eye to detect the discontinuity at the knots.
We now need X values that we can use for prediction purposes. In the code below we first find the range of the “inv2” variable. We then create a grid that includes all the possible values of “inv2” in increments of 1. Lastly, we use the “predict” function to develop the prediction model. We set the “se” argument to true as we will need this information. The code is below.
inv2lims<-range(Clothing$inv2)
inv2.grid<-seq(from=inv2lims[1],to=inv2lims[2])
pred<-predict(fit,newdata=list(inv2=inv2.grid),se=T)We are now ready to plot our model. The code below graphs the model and includes the regression line (red), confidence interval (green), as well as the location of each knot (blue)
plot(Clothing$inv2,Clothing$tsales,main="Regression Spline Plot")
lines(inv2.grid,pred$fit,col='red',lwd=3)
lines(inv2.grid,pred$fit+2*pred$se.fit,lty="dashed",lwd=2,col='green')
lines(inv2.grid,pred$fit-2*pred$se.fit,lty="dashed",lwd=2,col='green')
segments(12000,0,x1=12000,y1=5000000,col='blue' )
segments(60000,0,x1=60000,y1=5000000,col='blue' )
segments(150000,0,x1=150000,y1=5000000,col='blue' )
When this model was created it was essentially three models connected. Model on goes from the first blue line to the second. Model 2 goes form the second blue line to the third and model three was from the third blue line until the end. This kind of flexibility is valuable in understanding nonlinear relationship
Factors in R VIDEO
Factors in R
Logistic Polynomial Regression in R
Polynomial regression is used when you want to develop a regression model that is not linear. It is common to use this method when performing traditional least squares regression. However, it is also possible to use polynomial regression when the dependent variable is categorical. As such, in this post, we will go through an example of logistic polynomial regression.
Specifically, we will use the “Clothing” dataset from the “Ecdat” package. We will divide the “tsales” dependent variable into two categories to run the analysis. Below is the code to get started.
library(Ecdat)data(Clothing)There is little preparation for this example. Below is the code for the model
fitglm<-glm(I(tsales>900000)~poly(inv2,4),data=Clothing,family = binomial)Here is what we did
1. We created an object called “fitglm” to save our results
2. We used the “glm” function to process the model
3. We used the “I” function. This told R to process the information inside the parentheses as is. As such, we did not have to make a new variable in which we split the “tsales” variable. Simply, if sales were greater than 900000 it was code 1 and 0 if less than this amount.
4. Next, we set the information for the independent variable. We used the “poly” function. Inside this function, we placed the “inv2” variable and the highest order polynomial we want to explore.
5. We set the data to “Clothing”
6. Lastly, we set the “family” argument to “binomial” which is needed for logistic regression
Below is the results
summary(fitglm)##
## Call:
## glm(formula = I(tsales > 9e+05) ~ poly(inv2, 4), family = binomial,
## data = Clothing)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.5025 -0.8778 -0.8458 1.4534 1.5681
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 3.074 2.685 1.145 0.2523
## poly(inv2, 4)1 641.710 459.327 1.397 0.1624
## poly(inv2, 4)2 585.975 421.723 1.389 0.1647
## poly(inv2, 4)3 259.700 178.081 1.458 0.1448
## poly(inv2, 4)4 73.425 44.206 1.661 0.0967 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 521.57 on 399 degrees of freedom
## Residual deviance: 493.51 on 395 degrees of freedom
## AIC: 503.51
##
## Number of Fisher Scoring iterations: 13It appears that only the 4th-degree polynomial is significant and barely at that. We will now find the range of our independent variable “inv2” and make a grid from this information. Doing this will allow us to run our model using the full range of possible values for our independent variable.
inv2lims<-range(Clothing$inv2)
inv2.grid<-seq(from=inv2lims[1],to=inv2lims[2])The “inv2lims” object has two values. The lowest value in “inv2” and the highest value. These values serve as the highest and lowest values in our “inv2.grid” object. This means that we have values started at 350 and going to 400000 by 1 in a grid to be used as values for “inv2” in our prediction model. Below is our prediction model.
predsglm<-predict(fitglm,newdata=list(inv2=inv2.grid),se=T,type="response")Next, we need to calculate the probabilities that a given value of “inv2” predicts a store has “tsales” greater than 900000. The equation is as follows.
pfit<-exp(predsglm$fit)/(1+exp(predsglm$fit))Graphing this leads to interesting insights. Below is the code
plot(pfit)
You can see the curves in the line from the polynomial expression. As it appears. As inv2 increase the probability increase until the values fall between 125000 and 200000. This is interesting, to say the least.
We now need to plot the actual model. First, we need to calculate the confidence intervals. This is done with the code below.
se.bandsglm.logit<-cbind(predsglm$fit+2*predsglm$se.fit,predsglm$fit-2*predsglm$se.fit)
se.bandsglm<-exp(se.bandsglm.logit)/(1+exp(se.bandsglm.logit))The ’se.bandsglm” object contains the log odds of each example and the “se.bandsglm” has the probabilities. Now we plot the results
plot(Clothing$inv2,I(Clothing$tsales>900000),xlim=inv2lims,type='n')
points(jitter(Clothing$inv2),I((Clothing$tsales>900000)),cex=2,pch='|',col='darkgrey')
lines(inv2.grid,pfit,lwd=4)
matlines(inv2.grid,se.bandsglm,col="green",lty=6,lwd=6) In the code above we did the following.
In the code above we did the following.
1. We plotted our dependent and independent variables. However, we set the argument “type” to n which means nothing. This was done so we can add the information step-by-step.
2. We added the points. This was done using the “points” function. The “jitter” function just helps to spread the information out. The other arguments (cex, pch, col) our for aesthetics and our optional.
3. We add our logistic polynomial line based on our independent variable grid and the “pfit” object which has all of the predicted probabilities.
4. Last, we add the confidence intervals using the “matlines” function. Which includes the grid object as well as the “se.bandsglm” information.
You can see that these results are similar to when we only graphed the “pfit” information. However, we also add the confidence intervals. You can see the same dip around 125000-200000 were there is also a larger confidence interval. if you look at the plot you can see that there are fewer data points in this range which may be what is making the intervals wider.
Conclusion
Logistic polynomial regression allows the regression line to have more curves to it if it is necessary. This is useful for fitting data that is non-linear in nature.
Polynomial Regression in R
Polynomial regression is one of the easiest ways to fit a non-linear line to a data set. This is done through the use of higher order polynomials such as cubic, quadratic, etc to one or more predictor variables in a model.
Generally, polynomial regression is used for one predictor and one outcome variable. When there are several predictor variables it is more common to use generalized additive modeling/ In this post, we will use the “Clothing” dataset from the “Ecdat” package to predict total sales with the use of polynomial regression. Below is some initial code.
library(Ecdat)data(Clothing) str(Clothing)
## 'data.frame': 400 obs. of 13 variables:
## $ tsales : int 750000 1926395 1250000 694227 750000 400000 1300000 495340 1200000 495340 ...
## $ sales : num 4412 4281 4167 2670 15000 ...
## $ margin : num 41 39 40 40 44 41 39 28 41 37 ...
## $ nown : num 1 2 1 1 2 ...
## $ nfull : num 1 2 2 1 1.96 ...
## $ npart : num 1 3 2.22 1.28 1.28 ...
## $ naux : num 1.54 1.54 1.41 1.37 1.37 ...
## $ hoursw : int 76 192 114 100 104 72 161 80 158 87 ...
## $ hourspw: num 16.8 22.5 17.2 21.5 15.7 ...
## $ inv1 : num 17167 17167 292857 22207 22207 ...
## $ inv2 : num 27177 27177 71571 15000 10000 ...
## $ ssize : int 170 450 300 260 50 90 400 100 450 75 ...
## $ start : num 41 39 40 40 44 41 39 28 41 37 ...We are going to use the “inv2” variable as our predictor. This variable measures the investment in automation by a particular store. We will now run our polynomial regression model.
fit<-lm(tsales~poly(inv2,5),data = Clothing)
summary(fit)##
## Call:
## lm(formula = tsales ~ poly(inv2, 5), data = Clothing)
##
## Residuals:
## Min 1Q Median 3Q Max
## -946668 -336447 -96763 184927 3599267
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 833584 28489 29.259 < 2e-16 ***
## poly(inv2, 5)1 2391309 569789 4.197 3.35e-05 ***
## poly(inv2, 5)2 -665063 569789 -1.167 0.2438
## poly(inv2, 5)3 49793 569789 0.087 0.9304
## poly(inv2, 5)4 1279190 569789 2.245 0.0253 *
## poly(inv2, 5)5 -341189 569789 -0.599 0.5497
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 569800 on 394 degrees of freedom
## Multiple R-squared: 0.05828, Adjusted R-squared: 0.04633
## F-statistic: 4.876 on 5 and 394 DF, p-value: 0.0002428The code above should be mostly familiar. We use the “lm” function as normal for regression. However, we then used the “poly” function on the “inv2” variable. What this does is runs our model 5 times (5 is the number next to “inv2”). Each time a different polynomial is used from 1 (no polynomial) to 5 (5th order polynomial). The results indicate that the 4th-degree polynomial is significant.
We now will prepare a visual of the results but first, there are several things we need to prepare. First, we want to find what the range of our predictor variable “inv2” is and we will save this information in a grade. The code is below.
inv2lims<-range(Clothing$inv2)Second, we need to create a grid that has all the possible values of “inv2” from the lowest to the highest broken up in intervals of one. Below is the code.
inv2.grid<-seq(from=inv2lims[1],to=inv2lims[2])We now have a dataset with almost 400000 data points in the “inv2.grid” object through this approach. We will now use these values to predict “tsales.” We also want the standard errors so we se “se” to TRUE
preds<-predict(fit,newdata=list(inv2=inv2.grid),se=TRUE)We now need to find the confidence interval for our regression line. This is done by making a dataframe that takes the predicted fit adds or subtracts 2 and multiples this number by the standard error as shown below.
se.bands<-cbind(preds$fit+2*preds$se.fit,preds$fit-2*preds$se.fit)With these steps completed, we are ready to create our civilization.
To make our visual, we use the “plot” function on the predictor and outcome. Doing this gives us a plot without a regression line. We then use the “lines” function to add the polynomial regression line, however, this line is based on the “inv2.grid” object (40,000 observations) and our predictions. Lastly, we use the “matlines” function to add the confidence intervals we found and stored in the “se.bands” object.
plot(Clothing$inv2,Clothing$tsales)
lines(inv2.grid,preds$fit,lwd=4,col='blue')
matlines(inv2.grid,se.bands,lwd = 4,col = "yellow",lty=4)
Conclusion
You can clearly see the curvature of the line. Which helped to improve model fit. Now any of you can tell that we are fitting this line to mostly outliers. This is one reason we the standard error gets wider and wider it is because there are fewer and fewer observations on which to base it. However, for demonstration purposes, this is a clear example of the power of polynomial regression.
















