This is a fork of bonito which integrates slow5. It currently works for the basecaller module, using seq_reads_multi() from the pyslow5 library.
Random access can easily be added with get_read() and get_read_list_multi() for the other bonito modules.
A few flags are added as well:
- "--slow5", action="store_true", default=False: expect slow5/blow5 files rather than fast5 for input
- "--slow5_threads", default=4 : number of threads to use in pyslow5 (threads handled on C side of library)
- "--procs", default=8 : number of processors to use in bonito (have not tested altering these, and kept default for testing)
- "--slow5_batchsize", default=4096 : batch size to use for memory management with slow5. Larger batchsize uses more RAM
python3.8 -m venv bonitoS5
source bonitoS5/bin/activate
pip install --upgrade pip
git clone https://github.com/Psy-Fer/bonito.git
cd bonito
pip install -r requirements.txt
python setup.py develop
bonito --helpBonito results will show that for a single sample BLOW5 and FAST5 give similar runtime as FAST5 multiprocessing is implemented. However, we show that FAST5 multiprocessing is not scalable beyond a few samples as random accesses quickly saturate the disk system capability, whereas sequential I/O in BLOW5 is very lightweight and allows scalability over dozens of samples.
When the disk system is solely allocated for Bonito: FAST5: 31.5 min SLOW5: 28 min
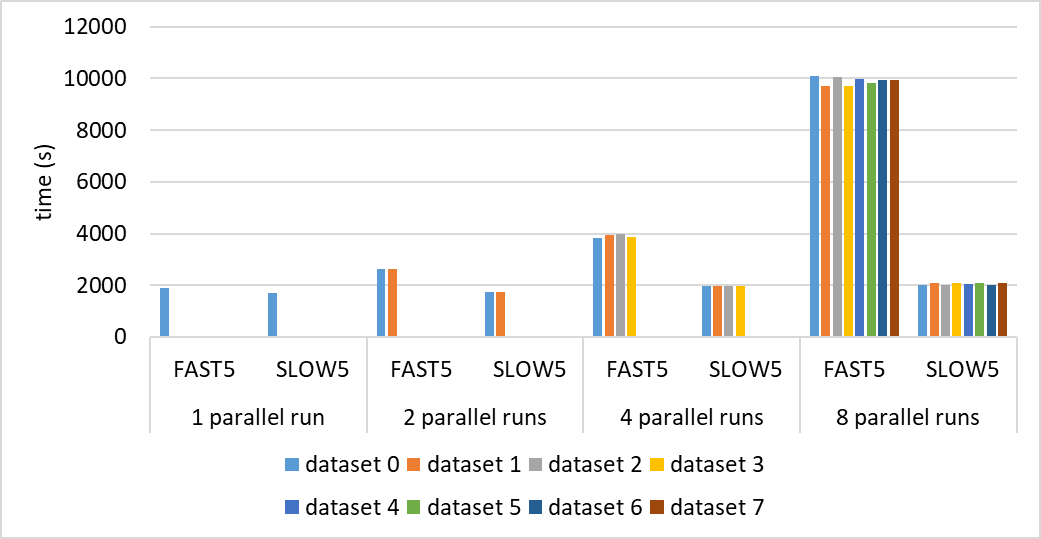
On a GPU server with 4 Tesla V100s and 40 cores reading data from a mounted NAS, we executed four instances of FAST5 basecalling and SLOW5 basecalling in parallel. FAST5 instances took around 1 hour whereas SLOW5 instances took 30 mins. This shows that SLOW5 is lightweight and can scale up.
FAST5 times: 1:03:40 1:05:44 1:06:02 1:04:40 SLOW5 times: 32:39.06 32:59.78 32:40.15 32:51.6
When the disk system is under extreme load (represents a scenario where we basecall hundreds of samples on an local cluster). Simulated scenario by running fio to introduce a huge disk I/O load. FAST5: 15 hours SLOW5: 52 min
Commands used for benchmarking
(yes, mem cache was cleared between all benchmarks)
bonito basecaller --batchsize 2048 -v dna_r9.4.1_e8.1_fast@v3.4 fast5/NA12878_SRE/20201027_0537_3A_PAF25452_97d631c6/fast5_pass/ --device cuda > a.fastq
bonito basecaller --batchsize 2048 -v dna_r9.4.1_e8.1_fast@v3.4 slow5_merged/ --slow5 --slow5_threads 8 --slow5_batchsize 4096 --device cuda > b.fastq
# synthetic workload to represent basecalling hundreds of samples in parallel:
fio --randrepeat=1 --ioengine=libaio --direct=1 --gtod_reduce=1 --name=test --filename=test --bs=100k --iodepth=32 --size=16G --readwrite=randreadThis figure with up to 8 parallel runs shows how slow5 scales better than fast5.
By increasing the speed by which a basecaller can process multiple parallel jobs, we can save users time, and if running on cloud infrastructure, a lot of money. This also shows how slow5 could increase the number of flowcells the basecallers could "keep up" with. (see also our guppy modifications and buttery-eel)
A PyTorch Basecaller for Oxford Nanopore Reads.
$ pip install --upgrade pip
$ pip install ont-bonito
$ bonito basecaller dna_r10.4_e8.1_sup@v3.4 /data/reads > basecalls.bamBonito supports writing aligned/unaligned {fastq, sam, bam, cram}.
$ bonito basecaller dna_r10.4_e8.1_sup@v3.4 --reference reference.mmi /data/reads > basecalls.bamBonito will download and cache the basecalling model automatically on first use but all models can be downloaded with -
$ bonito download --models --show # show all available models
$ bonito download --models # download all available modelsThe default ont-bonito package is built against CUDA 10.2 however CUDA 11.1 and 11.3 builds are available.
$ pip install -f https://download.pytorch.org/whl/torch_stable.html ont-bonito-cuda111Modified base calling is handled by Remora.
$ bonito basecaller dna_r10.4_e8.1_sup@v3.4 /data/reads --modified-bases 5mC --reference ref.mmi > basecalls_with_mods.bamSee available modified base models with the remora model list_pretrained command.
To train a model using your own reads, first basecall the reads with the additional --save-ctc flag and use the output directory as the input directory for training.
$ bonito basecaller dna_r9.4.1 --save-ctc --reference reference.mmi /data/reads > /data/training/ctc-data/basecalls.sam
$ bonito train --directory /data/training/ctc-data /data/training/model-dirIn addition to training a new model from scratch you can also easily fine tune one of the pretrained models.
bonito train --epochs 1 --lr 5e-4 --pretrained dna_r10.4_e8.1_sup@v3.4 --directory /data/training/ctc-data /data/training/fine-tuned-modelIf you are interested in method development and don't have you own set of reads then a pre-prepared set is provide.
$ bonito download --training
$ bonito train /data/training/model-dirAll training calls use Automatic Mixed Precision to speed up training. To disable this, set the --no-amp flag to True.
$ git clone https://github.com/nanoporetech/bonito.git # or fork first and clone that
$ cd bonito
$ python3 -m venv venv3
$ source venv3/bin/activate
(venv3) $ pip install --upgrade pip
(venv3) $ pip install -r requirements.txt
(venv3) $ python setup.py developbonito view- view a model architecture for a given.tomlfile and the number of parameters in the network.bonito train- train a bonito model.bonito evaluate- evaluate a model performance.bonito download- download pretrained models and training datasets.bonito basecaller- basecaller (.fast5->.bam).
- Sequence Modeling With CTC
- Quartznet: Deep Automatic Speech Recognition With 1D Time-Channel Separable Convolutions
- Pair consensus decoding improves accuracy of neural network basecallers for nanopore sequencing
(c) 2019 Oxford Nanopore Technologies Ltd.
Bonito is distributed under the terms of the Oxford Nanopore Technologies, Ltd. Public License, v. 1.0. If a copy of the License was not distributed with this file, You can obtain one at http://nanoporetech.com
Research releases are provided as technology demonstrators to provide early access to features or stimulate Community development of tools. Support for this software will be minimal and is only provided directly by the developers. Feature requests, improvements, and discussions are welcome and can be implemented by forking and pull requests. However much as we would like to rectify every issue and piece of feedback users may have, the developers may have limited resource for support of this software. Research releases may be unstable and subject to rapid iteration by Oxford Nanopore Technologies.