Cracking the Code of Food Intolerance Reports with AI
From Idea to Innovation: The Technical Journey of Health Cleo
Day 1: The Spark on Jeju Island
It all started on a bustling street in Jeju, South Korea. I stood there, overwhelmed by the vibrant array of street food, but paralyzed by uncertainty. As someone with multiple food intolerances, I found myself in a familiar predicament: how could I safely navigate this culinary paradise without risking my health?

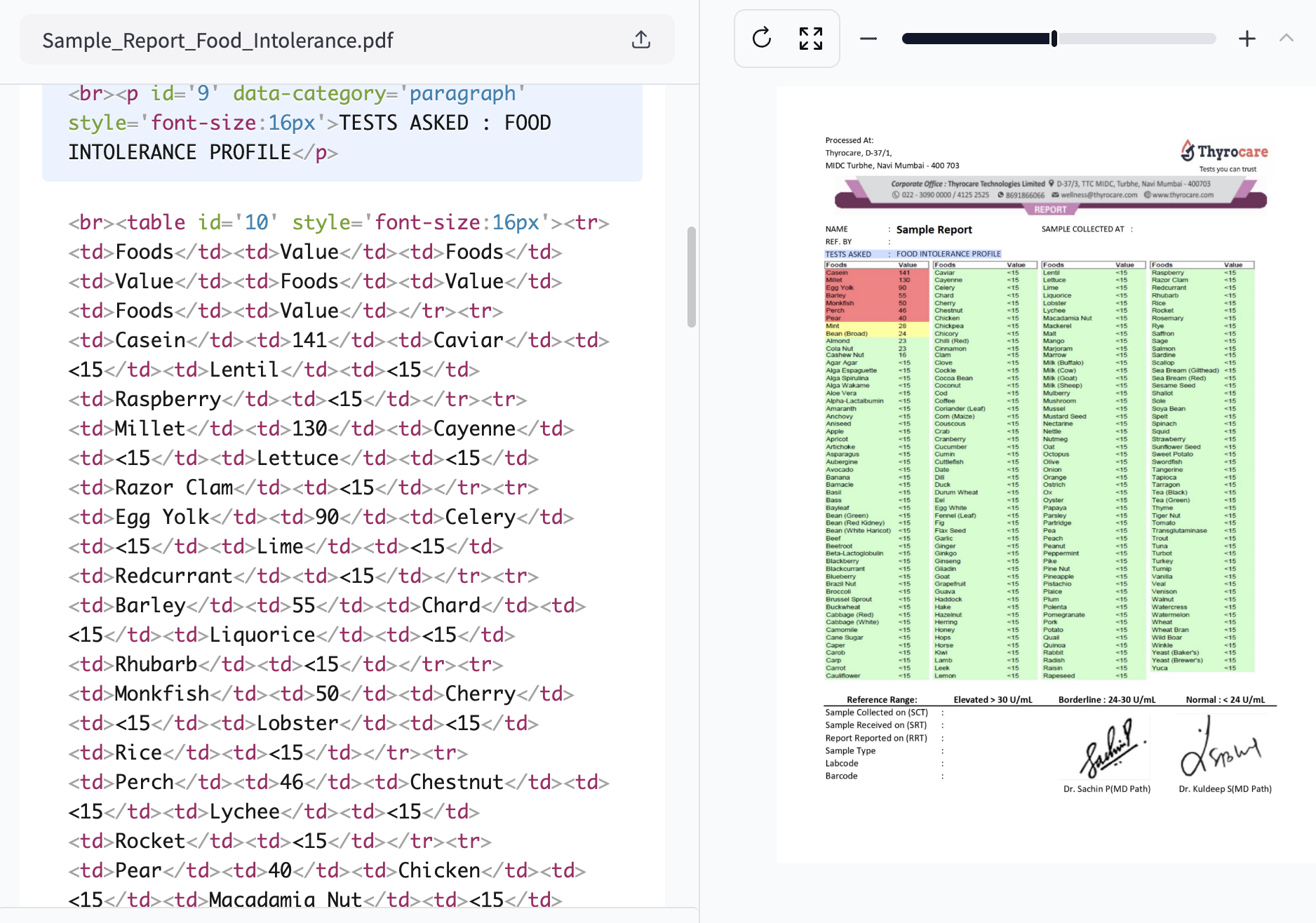
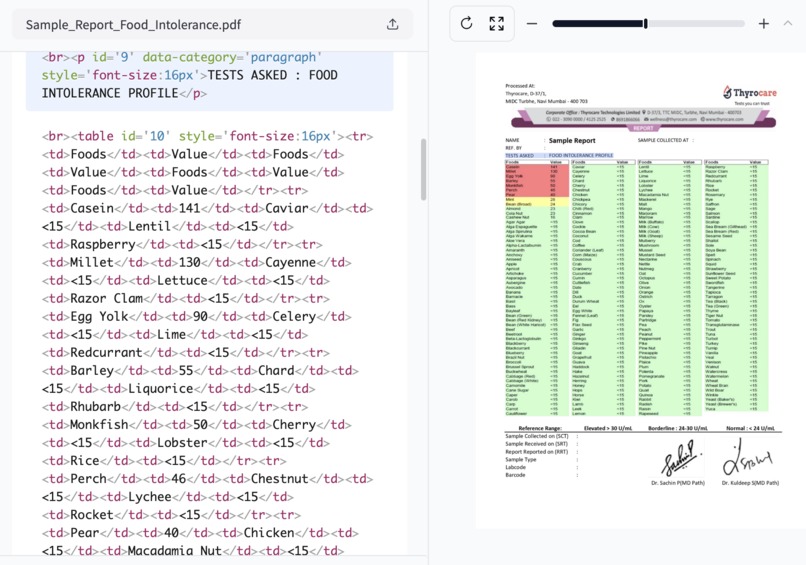
I pulled out my food intolerance report, a crumpled piece of paper filled with numbers and medical jargon. As I squinted at the tiny print, trying to remember which foods were safe and which weren't, a thought struck me: why isn't there an app for this?
Day 2: Research and Problem Definition
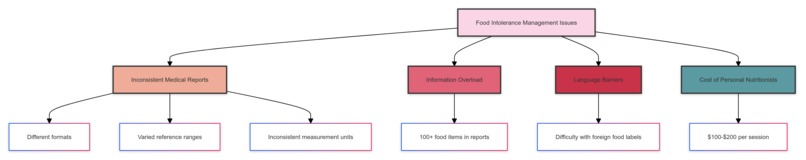
Back home, I dove into research. I discovered I wasn't alone in my struggles:
- Inconsistent Medical Reports: Each lab uses different formats, reference ranges, and measurement units.
- Information Overload: The average food intolerance report contains data on 100+ food items.
- Language Barriers: For travellers, deciphering foreign food labels adds another layer of complexity.
- Cost: Personal nutritionists are expensive, costing an average of USD 100 - USD 200 per session.
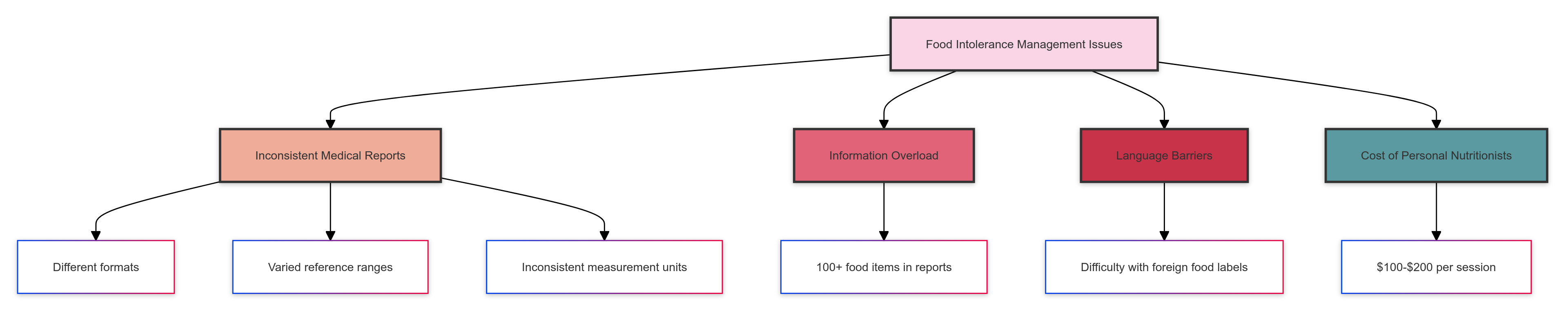
The problem was clear: we needed a solution to interpret varied medical reports, remember complex dietary restrictions, and provide real-time food safety advice.
Day 3: Conceptualizing the Solution
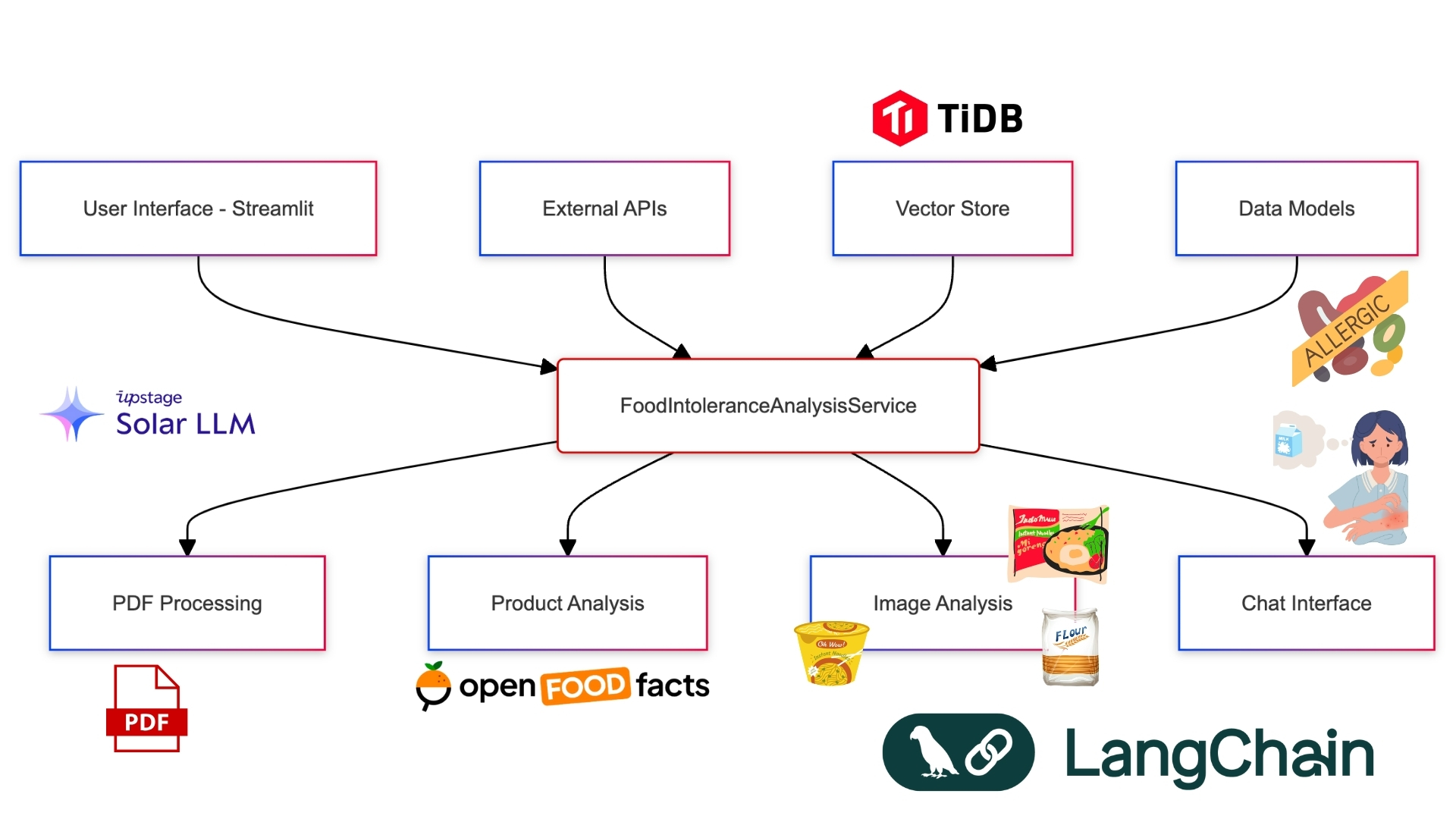
I envisioned an AI-powered app that could:
- Parse any food intolerance report
- Create a personalized dietary profile
- Analyze food products in real-time
- Provide safe eating recommendations
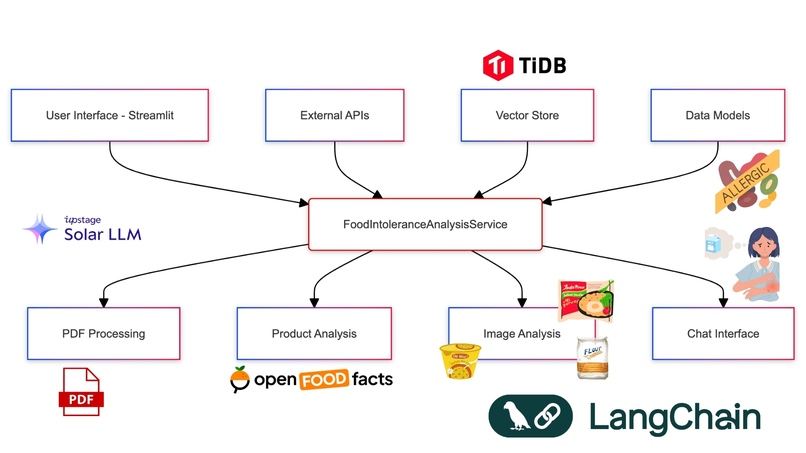
But how to build it? I needed a powerful AI backbone that could handle complex language processing, image recognition, and data analysis. After evaluating various options, I discovered Upstage AI's suite of APIs and TiDB vector db. Their combination of document parsing, OCR, embedding, and language models with vector db seemed perfect for our needs.
Day 4: Laying the Groundwork
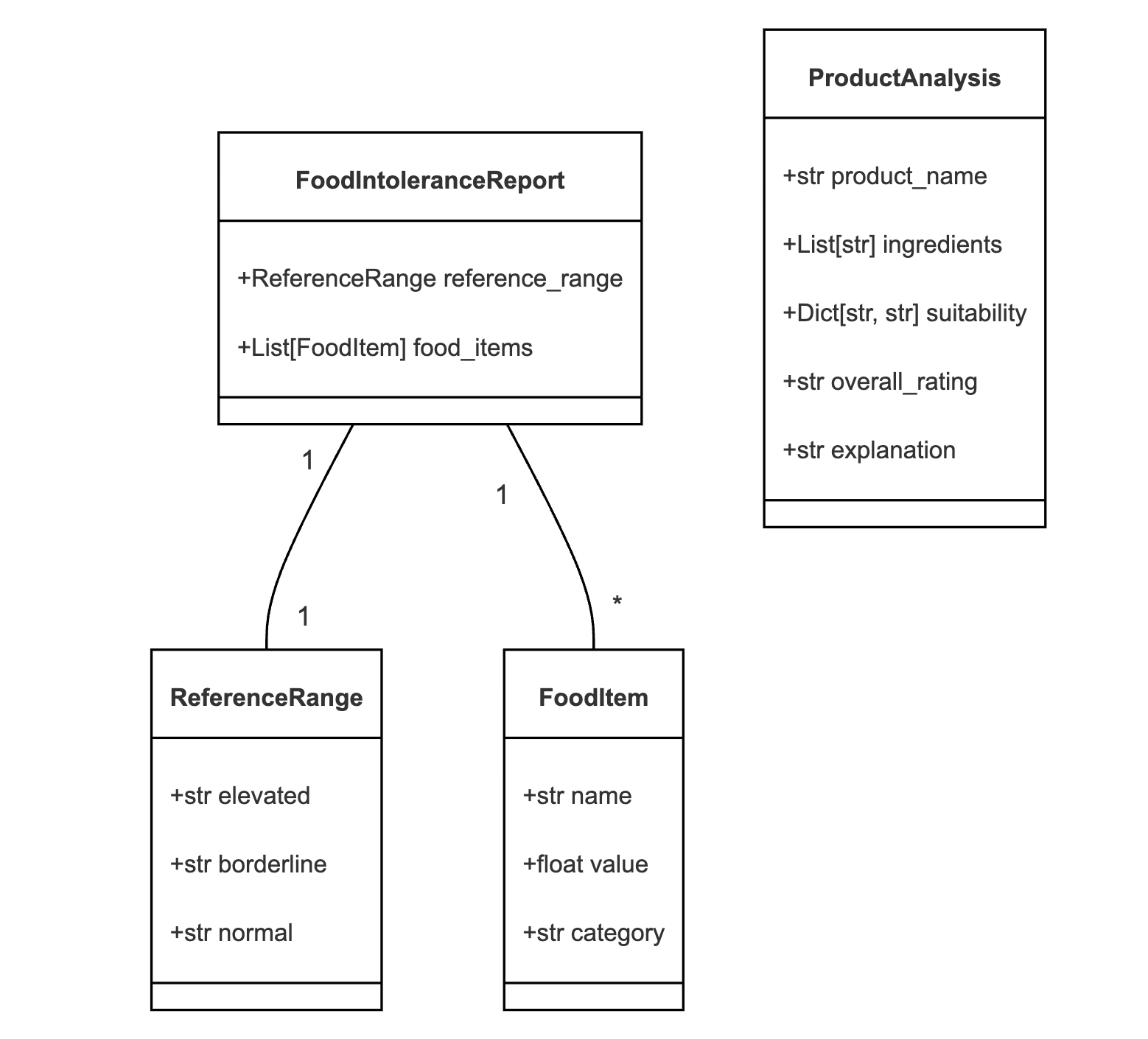
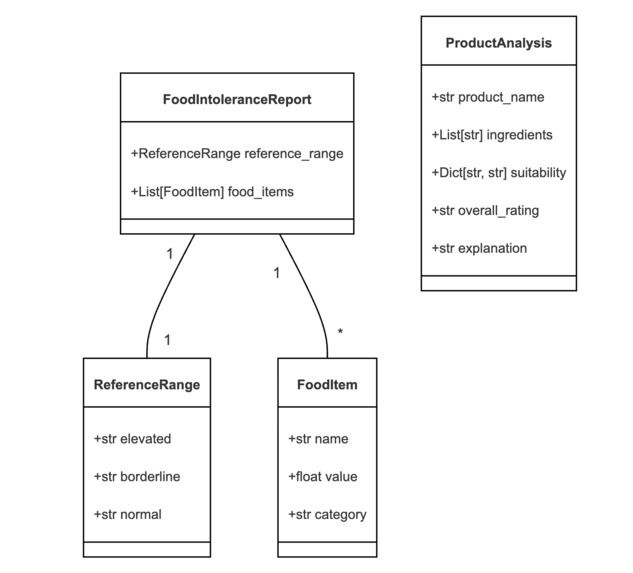
With the concept clear and the tools chosen, it was time to start coding. I began by setting up the project structure and defining our data models using Pydantic:
from pydantic import BaseModel
from typing import List, Dict
class ReferenceRange(BaseModel):
elevated: str
borderline: str
normal: str
class FoodItem(BaseModel):
name: str
value: float
category: str
class FoodIntoleranceReport(BaseModel):
reference_range: ReferenceRange
food_items: List[FoodItem]
class ProductAnalysis(BaseModel):
product_name: str
ingredients: List[str]
suitability: Dict[str, str]
overall_rating: str
explanation: str
Using Pydantic not only ensured type safety but also served as self-documenting code, clearly defining the structure of our data.
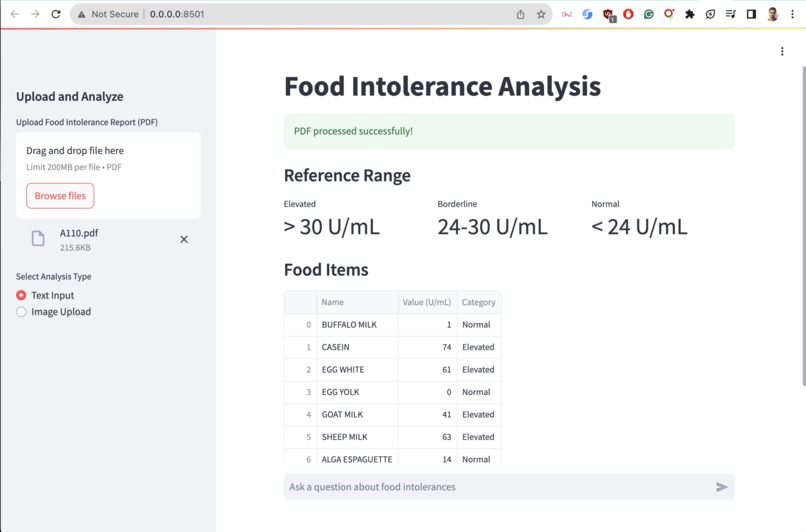
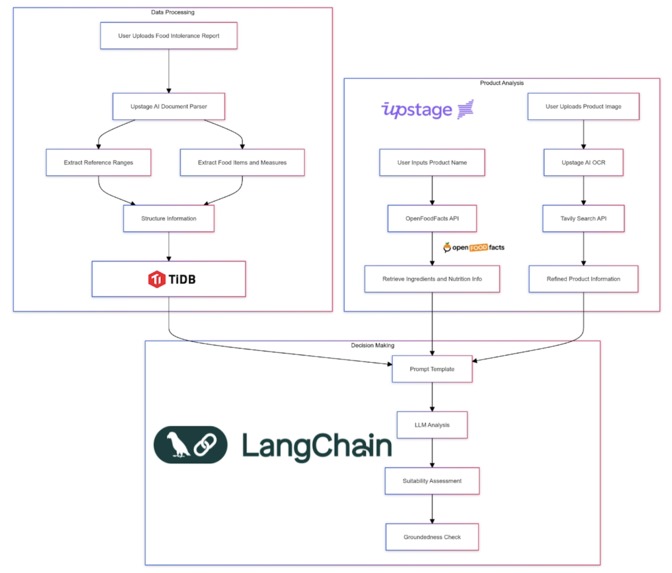
Day 5: Tackling Report Inconsistency
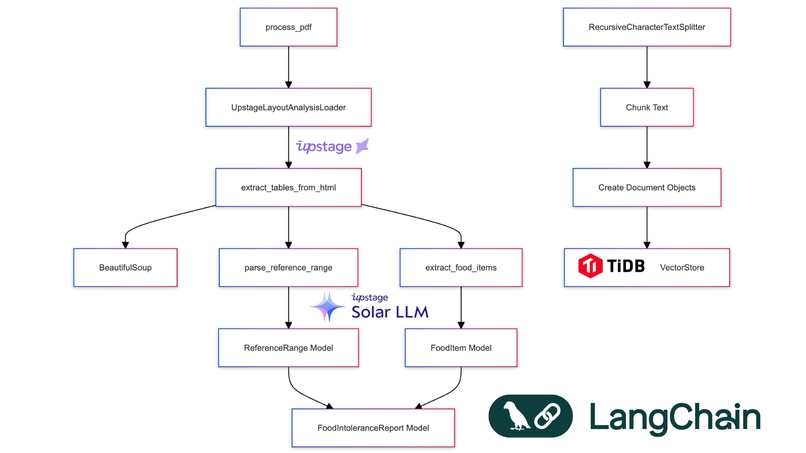
The first major challenge was parsing inconsistent medical reports. I leveraged Upstage's document parser and OCR capabilities, combined with custom HTML parsing and prompt engineering:
TiDB Integration: Powering Scalable Vector Storage
Health Cleo leverages TiDB, a distributed SQL database, to provide scalable and efficient vector storage for our food intolerance data. This integration allows us to handle large volumes of data while maintaining quick retrieval times, crucial for real-time product analysis.
Setting Up TiDB Connection
We initialize the TiDB connection in our FoodIntoleranceAnalysisService class:
from langchain_community.vectorstores import TiDBVectorStore
from sqlalchemy import Column, Integer, MetaData, String, Table, create_engine, JSON
class FoodIntoleranceAnalysisService:
def __init__(self, upstage_api_key: str, tavily_api_key: str, tidb_connection_string: str):
# ... other initializations ...
self.tidb_connection_string = tidb_connection_string
self.vector_store = self._initialize_vector_store()
def _initialize_vector_store(self):
return TiDBVectorStore(
connection_string=self.tidb_connection_string,
embedding_function=self.embeddings,
table_name="upstage-tidb-store",
distance_strategy="cosine"
)
This setup allows us to use TiDB as our vector store, leveraging its distributed nature for efficient storage and retrieval of embedding vectors.
Storing Extracted Data
When processing PDF reports, we now store the extracted tables in TiDB:
def process_pdf(self, pdf_path: str) -> FoodIntoleranceReport:
# ... existing code for loading and processing PDF ...
chunked_tables = self.text_splitter.split_text("\n".join(all_tables))
# Store the extracted tables in TiDB
documents_to_insert = []
for chunk in chunked_tables:
documents_to_insert.append(
Document(
page_content=chunk,
metadata={
"source": pdf_path,
"chunk_type": "table"
}
)
)
self.vector_store.add_texts([doc.page_content for doc in documents_to_insert], [doc.metadata for doc in documents_to_insert])
# ... rest of the processing ...
This approach allows us to persistently store the extracted information, making it available for future queries without needing to reprocess the PDF.
Querying the Vector Store
We've updated our querying method to use the TiDB vector store:
def _query_vectorstore(self, query: str) -> str:
relevant_docs = self.vector_store.similarity_search_with_score(query, k=3)
context = "\n".join([doc.page_content for doc, _ in relevant_docs])
prompt = f"""
Based on the following context, answer the question:
Context:
{context}
Question: {query}
Answer:
"""
response = self.llm([HumanMessage(content=prompt)])
return response.content
This method performs a similarity search in the TiDB vector store, retrieves the most relevant documents, and uses them as context for the language model to answer queries.
Benefits of TiDB Integration
- Scalability: TiDB's distributed nature allows Health Cleo to handle growing volumes of food intolerance data efficiently.
- Performance: The vector indexing capabilities of TiDB enable fast similarity searches, crucial for real-time product analysis.
- Persistence: Storing processed data in TiDB allows for quick retrieval in subsequent sessions, improving the overall user experience.
- Consistency: As a transactional database, TiDB ensures that our data remains consistent even as we update and query it concurrently.
By integrating TiDB, Health Cleo gains a robust, scalable backend that can grow with the application's needs, ensuring fast and reliable access to food intolerance data as we expand our user base and feature set.
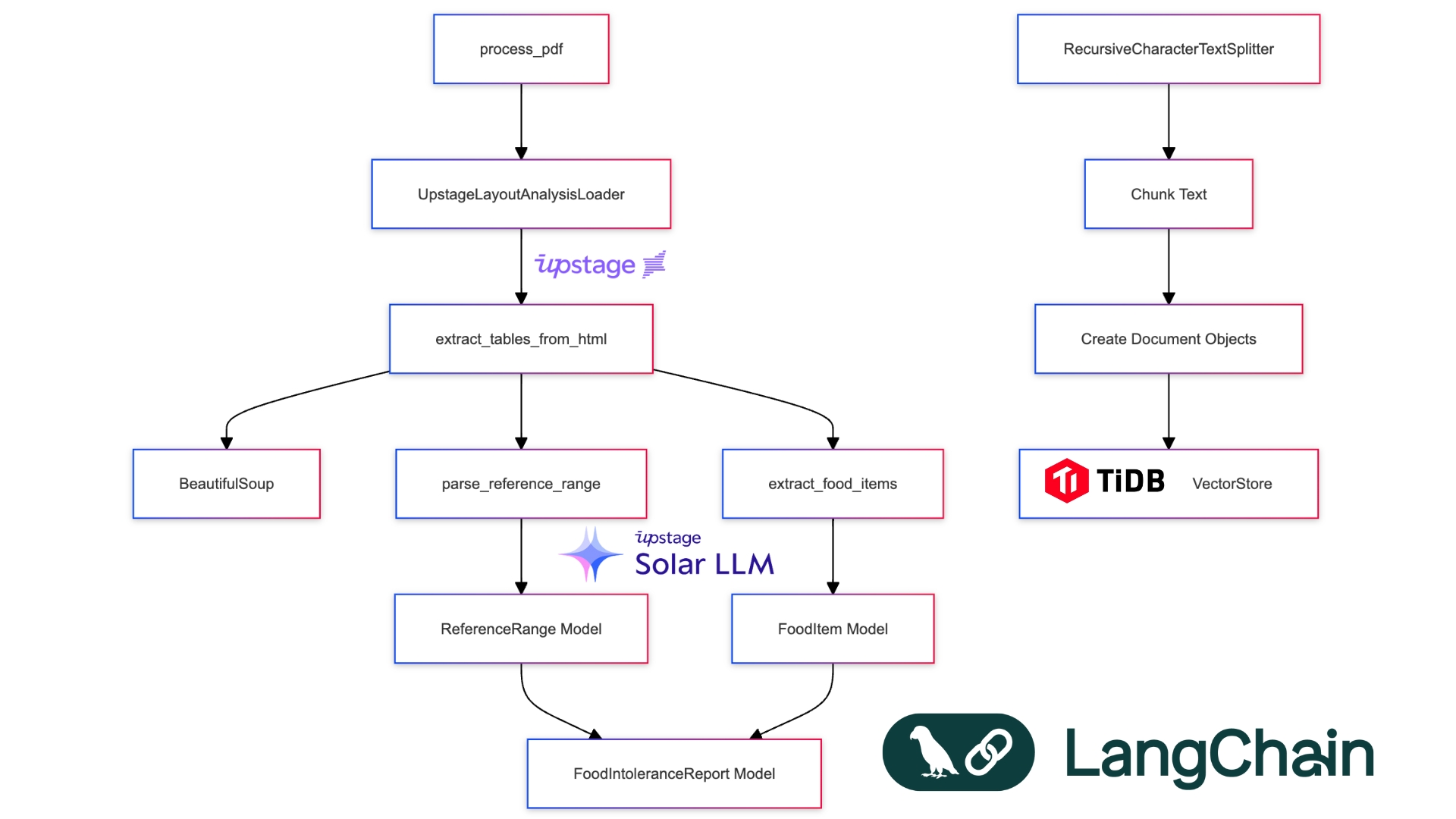
def extract_tables_from_html(html_content: str) -> List[str]:
soup = BeautifulSoup(html_content, 'html.parser')
return [str(table) for table in soup.find_all('table')]
def parse_reference_range(text: str) -> ReferenceRange:
elevated = re.search(r'Elevated[^\d]*(\d+(?:\.\d+)?)', text)
borderline = re.search(r'Borderline[^\d]*(\d+(?:\.\d+)?)[^\d]*(\d+(?:\.\d+)?)', text)
normal = re.search(r'Normal[^\d]*(\d+(?:\.\d+)?)', text)
return ReferenceRange(
elevated=f"> {elevated.group(1)} U/mL" if elevated else "",
borderline=f"{borderline.group(1)}-{borderline.group(2)} U/mL" if borderline else "",
normal=f"< {normal.group(1)} U/mL" if normal else ""
)
def categorize_food_item(value: float, reference_range: ReferenceRange) -> str:
elevated_threshold = float(re.search(r'\d+', reference_range.elevated).group())
borderline_range = [float(x) for x in re.findall(r'\d+', reference_range.borderline)]
if value >= elevated_threshold:
return "Elevated"
elif borderline_range[0] <= value <= borderline_range[1]:
return "Borderline"
else:
return "Normal"
Let's break down the key components:
HTML Parsing: The
extract_tables_from_htmlfunction uses BeautifulSoup to parse the HTML content and extract all table elements. This is crucial for handling the varied formats of medical reports.Reference Range Parsing: The
parse_reference_rangefunction uses regular expressions to extract the elevated, borderline, and normal ranges from the text. This flexibility allows us to handle different reporting formats.Food Item Categorization: The
categorize_food_itemfunction determines whether a food item's intolerance level is elevated, borderline, or normal based on the parsed reference range.PDF Processing: The
process_pdffunction ties everything together. It uses Upstage's LayoutAnalysisLoader to convert the PDF to HTML, extracts tables, chunks the content, and stores it in a TiDB vector database for efficient querying.
Output:
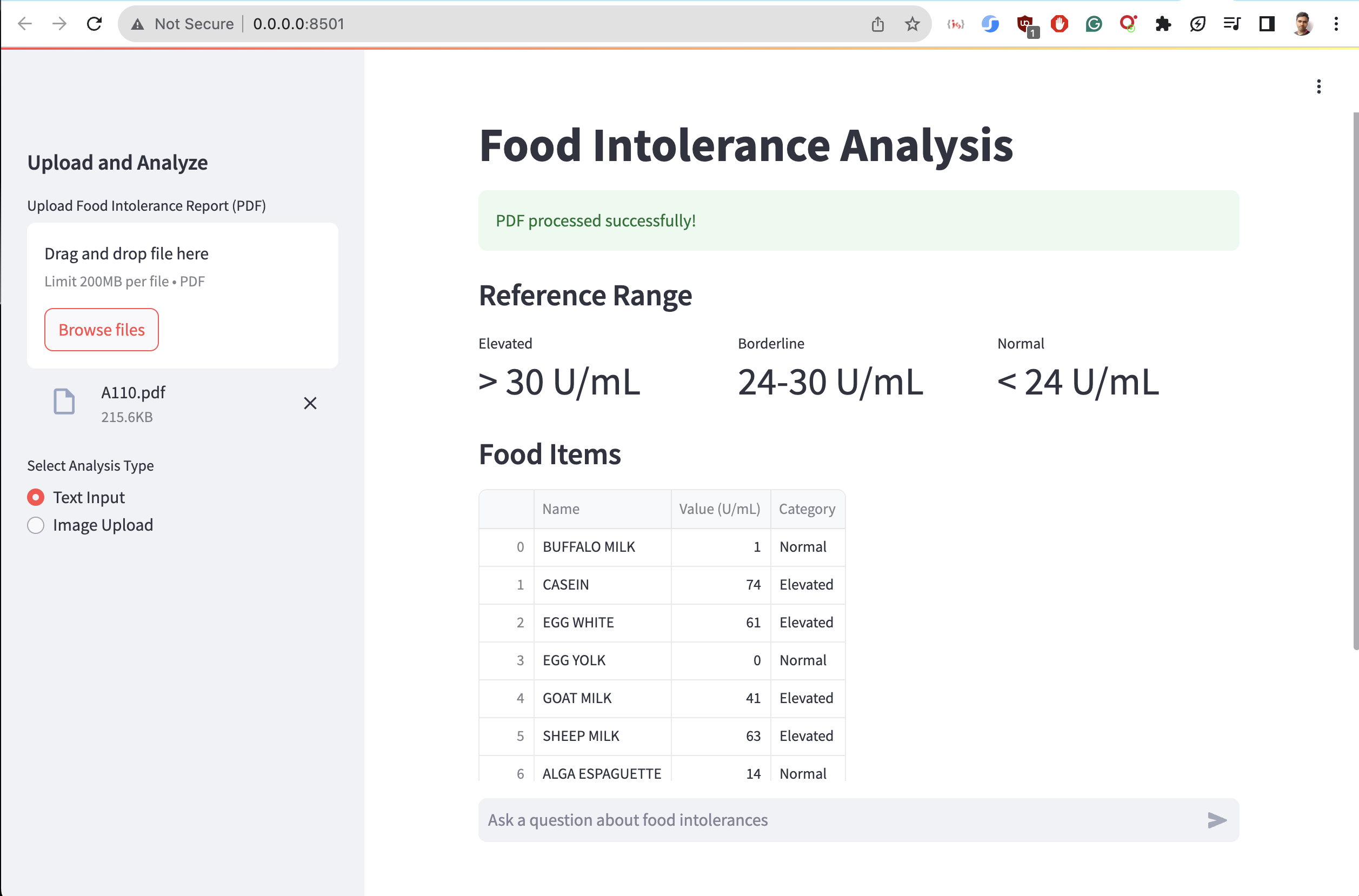
Day 6: Implementing Prompt Engineering and RetrievalQA
To extract meaningful information from the parsed reports, I implemented a sophisticated prompt engineering system using LangChain's PromptTemplate and RetrievalQA:
def _extract_reference_range(self, tables: List[str]) -> ReferenceRange:
reference_range_query = PromptTemplate(
input_variables=["tables"],
template="""
Analyze the following table content and extract the reference range for Elevated, Borderline, and Normal food intolerance levels:
{tables}
Provide the results in the following format:
Elevated: > X U/mL
Borderline: Y-Z U/mL
Normal: < W U/mL
"""
)
results = []
for chunk in tables:
result = self._query_vectorstore(reference_range_query.format(tables=chunk))
results.append(result)
combined_result = " ".join(results)
return parse_reference_range(combined_result)
def _query_vectorstore(self, query: str) -> str:
qa_chain = RetrievalQA.from_chain_type(
llm=self.llm,
chain_type="stuff",
retriever=self.vectorstore.as_retriever(search_kwargs={"k": 3}),
)
return qa_chain.run(query)
def _check_ingredient_suitability(self, ingredients: List[str]) -> Dict[str, str]:
suitability_query = PromptTemplate(
input_variables=["ingredients", "food_items"],
template="""
Based on the following food intolerance data:
{food_items}
Analyze the suitability of these ingredients:
{ingredients}
For each ingredient, classify it as:
- Suitable: if it's not in the list or has a "Normal" value
- Caution: if it's in the list with a "Borderline" value
- Avoid: if it's in the list with an "Elevated" value
- Unknown: if there's not enough information
Provide the result in the following format:
Ingredient: Classification
"""
)
# Split ingredients into chunks if necessary
ingredient_chunks = self.text_splitter.split_text(", ".join(ingredients))
food_items_text = "\n".join([f"{item.name}: {item.value} U/mL - {item.category}" for item in self.food_items])
all_suitability = {}
for chunk in ingredient_chunks:
result = self._query_vectorstore(suitability_query.format(
ingredients=chunk,
food_items=food_items_text
))
chunk_suitability = self._parse_suitability_result(result)
all_suitability.update(chunk_suitability)
return all_suitability
Here's what's happening:
Prompt Engineering: The
PromptTemplatein_extract_reference_rangecreates a structured query for the language model. This prompt engineering ensures that we get consistent, formatted responses regardless of the input variation.RetrievalQA: The
_query_vectorstorefunction sets up a RetrievalQA chain. This powerful feature combines the vector store (which contains our parsed report data) with the language model to answer complex queries about the report content.Chunked Processing: We process the tables in chunks, allowing us to handle large reports efficiently while maintaining context.
This approach allows us to extract structured information from unstructured or semi-structured medical reports, a key innovation in dealing with the inconsistency problem.
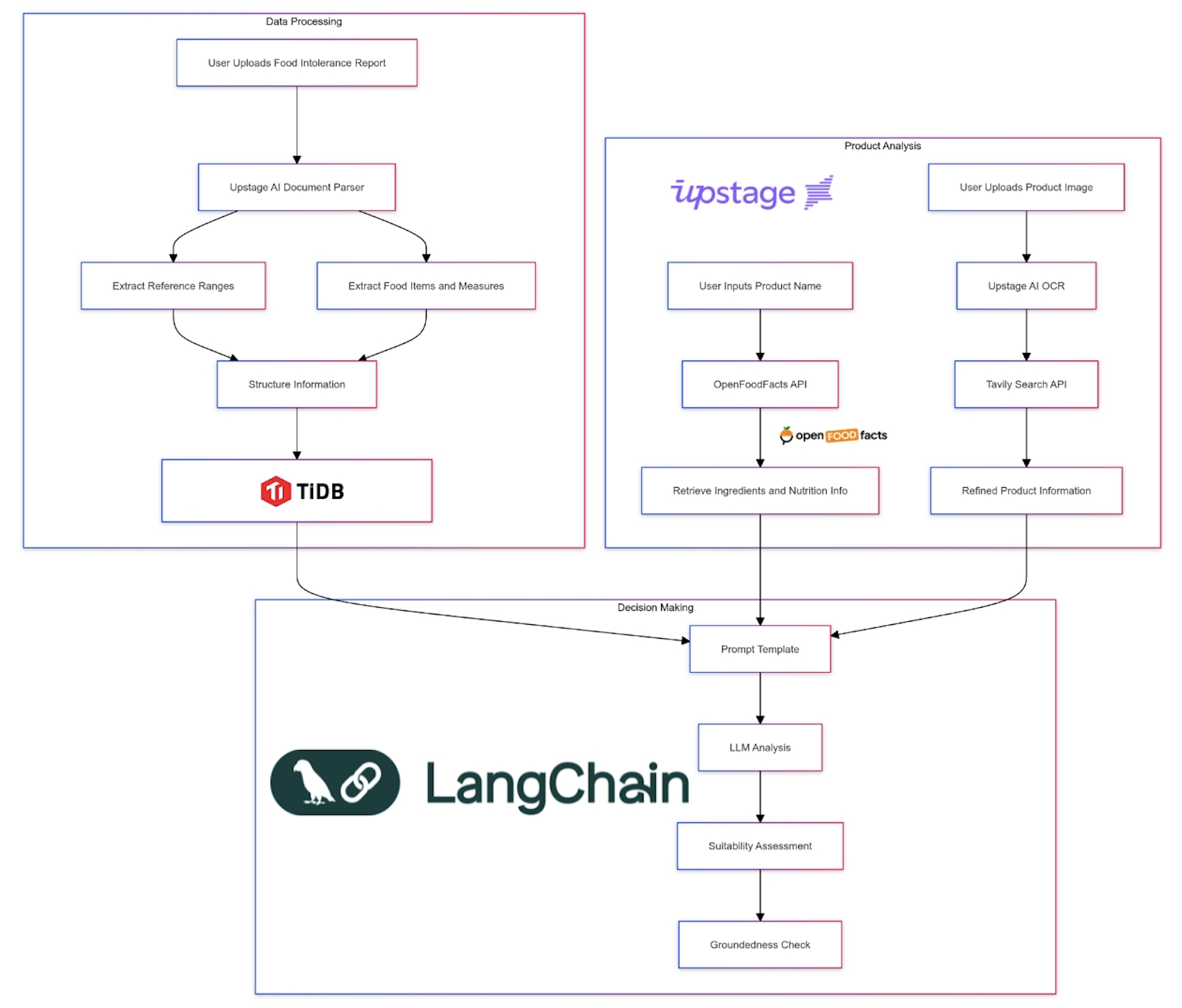
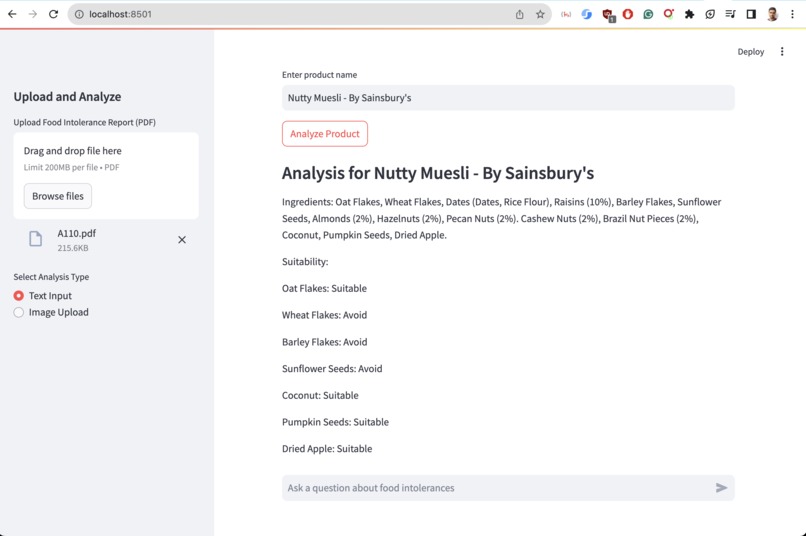
Day 6: Building the Product Analysis Pipeline
Next, I tackled the challenge of real-time product analysis. This required integrating multiple technologies:
def analyze_product_from_image(self, image_path: str) -> ProductAnalysis:
ocr_text = self.ocr_product_image(image_path)
context = self.tavily.search(query=f"Find product name for any such product that carries similar texts and brand '{ocr_text}'")
product_name_query = PromptTemplate(
input_variables=["context"],
template="Extract the most likely product name from the following context:\n\n{context}\n\nProduct Name:"
).format(context=context['results'][0]['content'])
product_name = self._query_vectorstore(product_name_query)
ingredients = self.get_product_ingredients(product_name)
return self.analyze_product(product_name, ingredients)
def get_product_ingredients(self, product_name: str) -> List[str]:
result = self.api.product.text_search(product_name, page=1, page_size=1)
if result['products']:
# Try to get the ingredients in English first
ingredients_text = result['products'][0].get('ingredients_text_en', '')
# If not found or empty, fall back to the default ingredients_text
if not ingredients_text:
ingredients_text = result['products'][0].get('ingredients_text', '')
# Return the list of ingredients, split by comma and stripped of extra spaces
return [ing.strip() for ing in ingredients_text.split(',') if ing.strip()]
return []
Output

This pipeline combines OCR, web search, language model querying, and our custom analysis logic to provide instant, accurate product assessments. Since OCR doesn't have contextual knowledge we use Tavily API to find accurate product name. Then use Upstage LLM to return idea candidat as product name.
Day 7: Ensuring Reliability with Groundedness Checks
To prevent AI hallucinations - a critical concern when dealing with health information - I implemented Upstage's groundedness check:
def _generate_overall_rating(self, suitability: Dict[str, str]) -> Tuple[str, str]:
# ... (previous code)
groundedness_check = UpstageGroundednessCheck(api_key=self.upstage_api_key)
request_input = {
"context": suitability_text,
"answer": explanation,
}
groundedness_response = groundedness_check.invoke(request_input)
if groundedness_response == "notGrounded":
explanation += " (Note: This explanation may not be fully grounded in the provided data.)"
elif groundedness_response == "notSure":
explanation += " (Note: The accuracy of this explanation could not be confirmed.)"
return rating, explanation
This ensures that every piece of advice given is firmly rooted in factual data.
Day 7: Optimization and Testing
The final weeks were spent optimizing performance and conducting thorough testing. I implemented efficient chunking strategies for handling large documents:
self.text_splitter = RecursiveCharacterTextSplitter(
chunk_size=3000,
chunk_overlap=200,
length_function=self.llm.get_num_tokens
)
This allows us to process large documents in manageable pieces while maintaining context.
The Result: Health Cleo
After an intense week, Health Cleo was born. It's not just an app; it's a personal AI nutritionist that:
- Understand any food intolerance report
- Provides real-time product analysis
- Offers multilingual support for global travellers
- Ensures reliability through groundedness checks
But more than that, it's a testament to the power of innovative engineering combined with cutting-edge AI. By leveraging Upstage AI's suite of APIs and implementing clever software design patterns, we've created a solution that turns the complex world of food intolerances into manageable, actionable information.
Running Streamlit demo
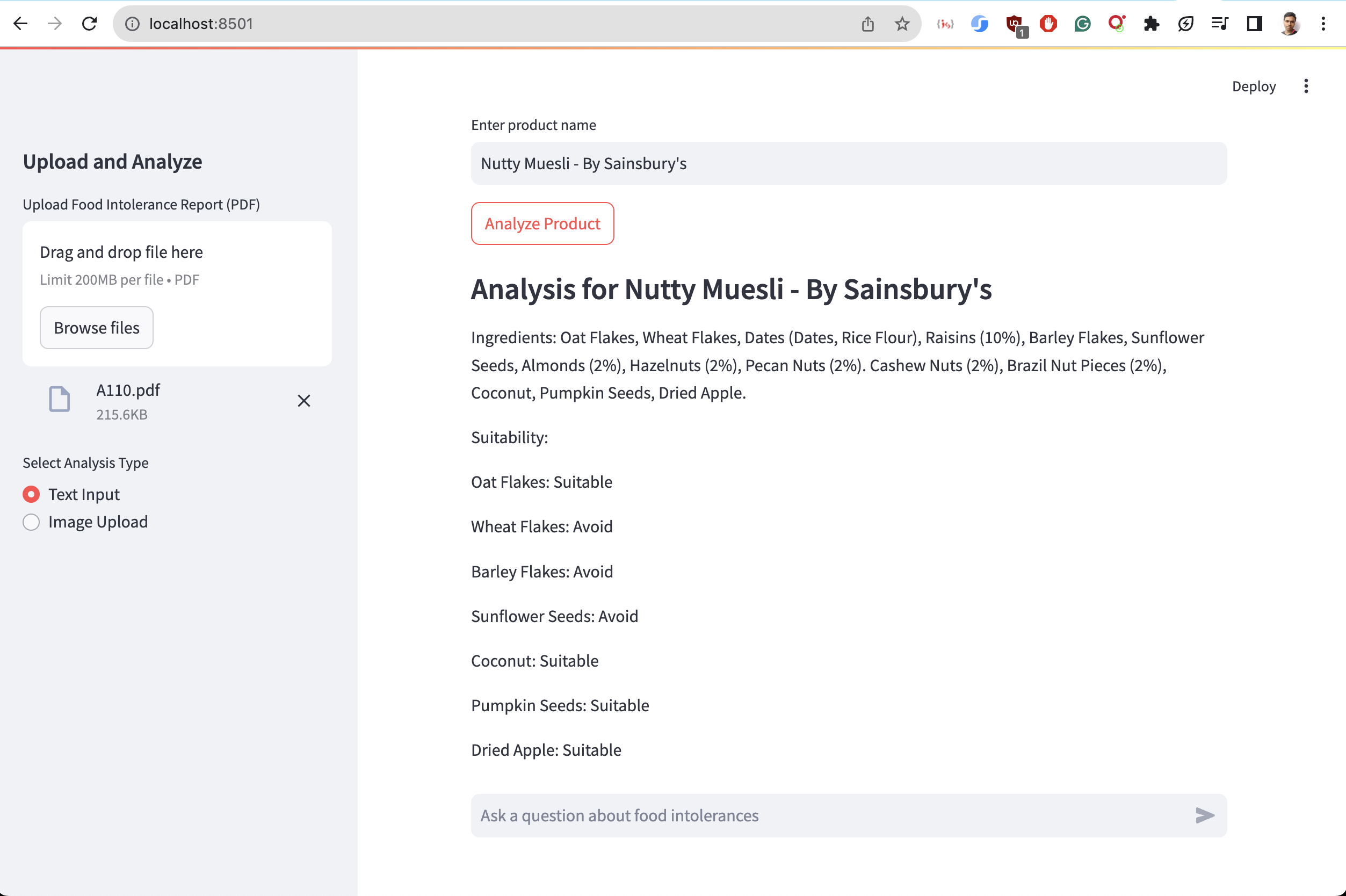
There is a run.sh script add you api keys and lauch the script in your terminal.
# make a new virtual env using python 3.11 (tested on MacOS 14.6.1)
# python3.11 -m venv .venv
# source .venv/bin/activate
# pip install -r requirements.txt
cd cleo
# put your API keys in app.py file beginning
streamlit run app.py --server.port 8501 --server.address 0.0.0.0
Looking Ahead
As I reflect on this journey, I'm excited about the future. There's so much more we can do:
- Integration with wearable devices like Rabbit R1, Humane AI pin for real-time capture, search and query
- Expansion of our food database to include more regional specialities
- Development of AR features for in-store shopping assistance

Health Cleo is just the beginning. We're working on integrating with wearable devices and expanding our database to include more regional and cultural specialities. Increasing other lab tests like allergy reports, etc. Imagine walking through a grocery store, and with a glance at a product through your AR glasses, you receive instant feedback on whether the product is safe for you to consume.
Here's to the future of personalized nutrition, powered by AI and driven by human ingenuity!
Built With
- langchain
- llm
- ocr
- python
- streamlit
- tidb
- upstage



Log in or sign up for Devpost to join the conversation.