Our project aims to leverage computer vision technology to make a contribution to neuroscience research. We were motivated to address the challenges in studying neurite growth dynamics in neurons, a fundamental aspect of neuroscience research. These cells play a crucial role in understanding neurodegeneration, neurodevelopmental disorders, neuron function, and differentiation, making them vital for advancing our understanding of diseases like Alzheimer's, Parkinson's, ASD, and more.
What we built
NeuraLIVE is a bioinformatic tool designed to streamline and enhance the process of analyzing experiment data on cultured neurons. It employs computer vision techniques to analyze brightfield microscopy images of these cells. Specifically, NeuraLIVE quantifies key aspects of neuron development:
- Cell Growth: NeuraLIVE determines whether the cells proliferate, a critical metric in various experiments.
- Cell Health: The program measures how healthy the cells are by comparing the changing cell shapes over time.
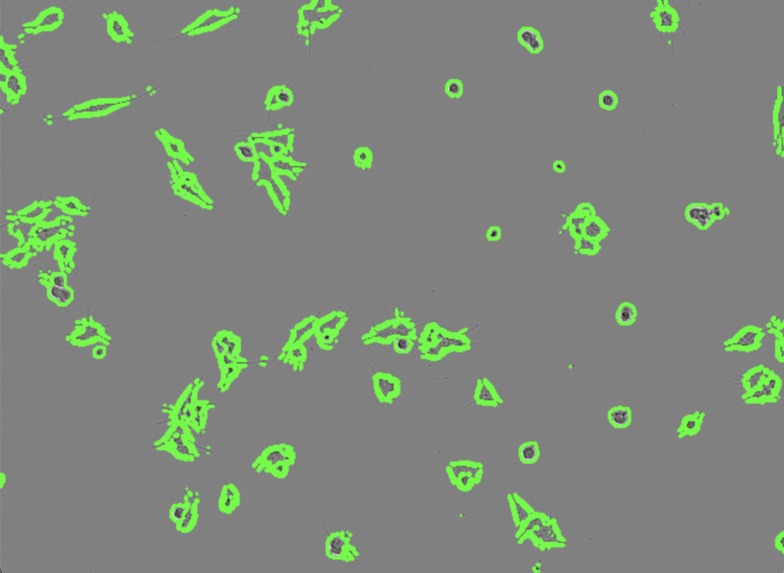
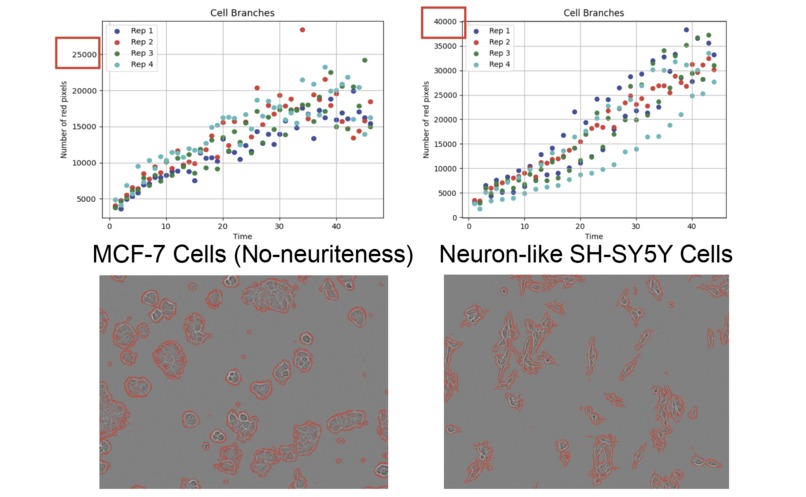
- Neuron Morphogenesis: It identifies sharpness in cell shape and thin, ridge-like objects that represent neurite growth and neuron morphogenesis-- essential processes for understanding the cell's ability to function as neurons.
We have developed an efficient and automated pipeline that can rapidly process and analyze large datasets of brightfield microscopy images. NeuraLIVE not only streamlines the analysis process but also allows analysis of experimental data in a high-throughput matter, truly integrating life science research with the booming field of big data analytics.
How we built it
We engineered NeuraLIVE by breaking it down into three fundamental tasks, each addressing a critical aspect of neuron analysis:
1: Neuron Identification - Utilizing Watershed Algorithm
The initial task is to analyze input images and determine the number of individual cells present. We implemented the Watershed Algorithm to segment and separate adjacent cell regions, which allows us to identify and quantify the cells in the images.
2: Neuron Classification - Identifying Round and Hypertrophic Cells
Task 2 was vital for distinguishing between round, unadhered cells, and spread-out, adhered cells. We employed contour plots and threshold filters to determine the shape of each cell in the images by distinguishing the number of edges in its contour. This classification not only allowed us to categorize cells but also provided valuable insights into their behavior. Round cells are often indicative of unadhered, unhealthy, cells, while spread-out cells signify adhesion and cell health. Finally, we used the numbers of the two types of cells to calculate the percentage of healthy cells in a given image.
3: Quantifying Neuriteness - Utilizing Ridge Detection Algorithm
The final task, Quantifying Neuriteness, addressed a crucial question: how much sharp, neurite-like structures are present in the cells? To accomplish this, we integrated the Ridge Detection Algorithm. This algorithm identifies and counts the number of straight, linear, and sharp objects within the images, which corresponded to neurogenesis. Quantification of neurite outgrowth is central to understanding neurodevelopment and neurodegenerative processes.
4: Front End Website To upload data sets and analyze the pictures we built a website using Flask as framework and HTML for the UI.
Challenges we ran into
Developing NeuraLIVE presented several challenges, including:
Data Parsing: We started off by taking all the images in the dataset as one culture and only found out later that they are actually from four different cultures. Thus, we had to restart the sorting process of the images to consider dividing the whole dataset into four cultures when plotting. This takes a while to implement and debug since the images are in random order, and parsing to find each image file's culture involves many unfamiliar syntax.
Ridge Detection: Quantifying neuriteness was extremely challenging--brightfield images are known for their difficulty in being analyzed, especially the sharper, thinner objects that do not reflect much light. In the beginning, the Ridge Detection Algorithm we implemented frequently identify non-neurite structures as neurites. We investigated the implementation of multiple ridge filters and explored options from various packages, and eventually choose the Meijuring filter with a customized greyscale threshold value to isolate the neurites from the cell bodies.
Accomplishments that we're proud of
Caroline: I'm most proud of cooperating so perfectly with my team in these 48 hours and finishing such an innovative project. Georges:_I am immensely proud of the technical innovation that NeuraLIVE represents. We successfully applied computer vision techniques to tackle a significant problem in neuroscience research. This project isn't just about code; it's about leveraging technology to advance our understanding of critical issues like neurodegeneration, neurodevelopmental disorders, and neuron function. The implication that NeuraLIVE can have on these fields is a testament to our dedication. _Millie: I am super proud of how supportive and collaborative we are to each other. Team work makes the dream work! Steve: As a student in both life science and computer science, it has always been my dream to carry out a project that combines my two fields of passion and make something that can potentially be useful to the research world. I believe NeuraLIVE, although being a product built within 48 hours of time, contains ideas and implementations that can be truly useful to the research field out there. I am so, so proud of my teammates, and thankful for theirs and our efforts in making this thing a reality along the way.
What we learned
Caroline: I was exposed to and learned a lot about the intersection of computer vision and biology through this project. Millie: I learned about neurons and many different useful python libraries for computer vision. George: As a student who knows much more about business than biology, this is something I haven't touched much before, but I absolutely love it! Hoping to maybe even take a biology class in the future. Steve: I learned so many useful image processing and computer vision libraries in Python, and learned about front-end development--something I have not worked with before!
What's next for NeuraLIVE
We plan to incorporate machine learning in future implementations, aiming to improve accuracy in object identification and segmentation. We also plan on optimizing several pixel-thresholding algorithm to improve the general runtime of our program.


Log in or sign up for Devpost to join the conversation.