PharmaNexus
Inspiration
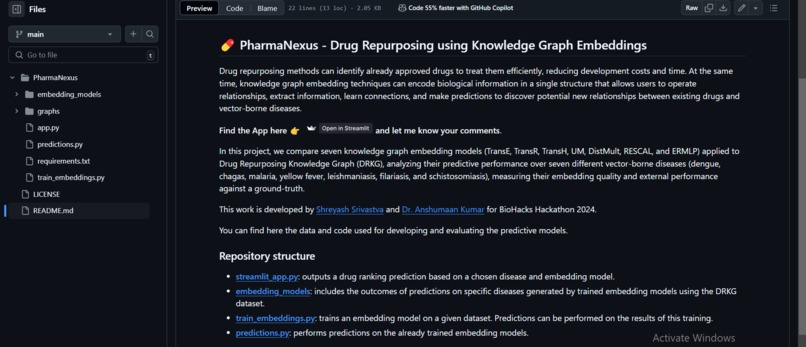
Drug repurposing has immense potential to revolutionize healthcare by finding new therapeutic uses for existing drugs, saving time, cost, and effort compared to de novo drug discovery. With the vast amount of biomedical data available today, we were inspired to leverage knowledge graphs and AI to unlock hidden connections and insights within this data, accelerating the discovery process. Our goal is to make drug repurposing more efficient and accessible, ultimately contributing to better patient outcomes.
What it does
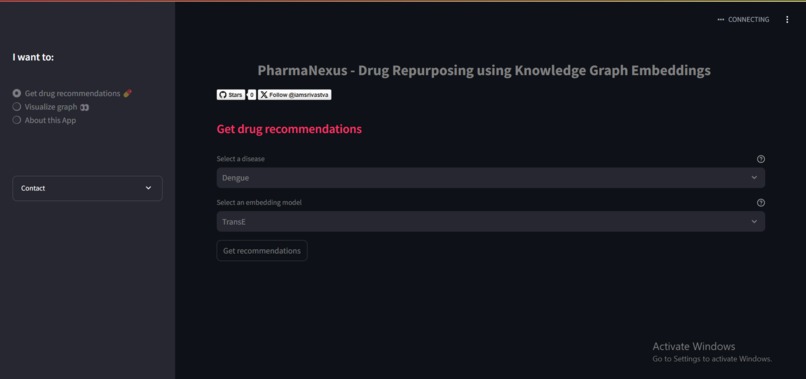
PharmaNexus is a cutting-edge platform that utilizes knowledge graphs to connect and analyze relationships between drugs, diseases, genes, and pathways. Key features include:
- Drug Discovery: Identifies potential drug candidates for repurposing.
- Relationship Visualization: Displays connections between biological entities for better insight.
- AI-Powered Predictions: Uses machine learning to rank and predict repurposing opportunities.
- Interactive Interface: Allows researchers to explore, filter, and validate findings intuitively.
How we built it
- Data Integration: Collected and preprocessed biomedical datasets from sources like DrugBank, PubMed, and clinical trial databases.
- Knowledge Graph Construction: Built a graph database using Neo4j to link drugs, diseases, genes, and biological pathways.
- AI Models: Applied graph neural networks and link prediction algorithms to discover potential repurposing candidates.
- Backend and API: Developed using Python and Flask for seamless data processing.
- Frontend: Built an intuitive Streamlit interface for researchers to interact with the platform.
Challenges we ran into
- Data Quality and Integration: Handling inconsistent and incomplete datasets required extensive cleaning and preprocessing.
- Scalability: Managing large-scale knowledge graphs while ensuring fast query response times was a technical challenge.
- Algorithm Optimization: Fine-tuning the graph neural networks to achieve meaningful and reliable predictions took significant effort.
- Interdisciplinary Knowledge: Bridging gaps between biology, data science, and AI required collaboration and research.
Accomplishments that we're proud of
- Successfully constructed a comprehensive and scalable biomedical knowledge graph.
- Developed a novel pipeline to identify drug repurposing candidates with scientific rigor.
- Designed an interactive and user-friendly platform to enable researchers to explore complex relationships easily.
- Demonstrated a real-world case study where PharmaNexus identified promising drug-disease associations.
What we learned
- The power of knowledge graphs in solving complex biomedical problems.
- Insights into biomedical data ecosystems and the importance of accurate annotations.
- Challenges and potential of applying AI techniques like graph neural networks in healthcare.
- The critical role of interdisciplinary collaboration in developing impactful solutions.
What's next for PharmaNexus
- Expand Data Coverage: Integrate more datasets like proteomics and metabolomics to enrich the knowledge graph.
- Enhance Predictions: Incorporate explainable AI models for better trust and transparency in predictions.
- Validation Pipeline: Collaborate with labs or researchers to experimentally validate high-potential candidates.
- Open Collaboration: Build a community around PharmaNexus, enabling researchers to contribute and share findings.
- Commercialization: Explore partnerships with pharmaceutical companies for real-world applications of the platform.
PharmaNexus aims to be at the forefront of AI-driven drug discovery, accelerating the development of treatments and improving lives globally.


Log in or sign up for Devpost to join the conversation.