Inspiration
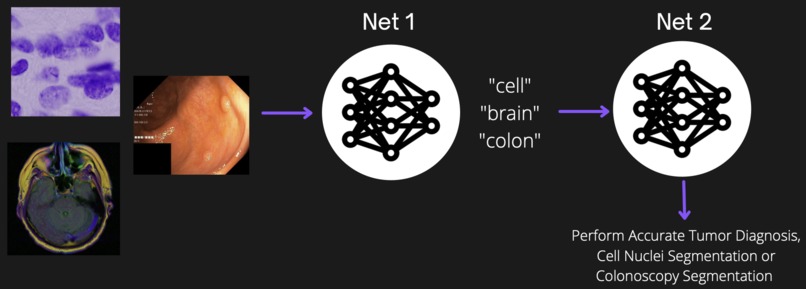
Neural networks are really good at really specific tasks. My submission focuses on the problem of continual learning: teaching neural networks new tasks without forgetting the old ones. During training, a neural network's parameters get finely tuned to a specific dataset, allowing it to provide accurate predictions on similar data. However, when that same neural network gets re-trained to learn a new task, its initial set of parameters are changed to fit this new data and as a result, it can no longer produce accurate results on the first task (it forgets how!) My project develops a model that is able to perform accurate brain tumor, cell nuclei, and colon tumor detection all on the same neural network by using a system that 'swaps' out its parameters rather than trying to re-use them.
What it does
My model implements two neural networks: the first net is a convolutional neural network (CNN) which detects the type of input image (as either "cell" "brain" or "colon") and the second net is a UNet which performs the medical segmentation. My second net uses the first net's output ("cell" "brain" or "colon") to determine which set of weights to 'swap' in before performing its segmentation. This way, the UNet can accurately perform different tasks by effectively learning which set of parameters (ie weights) to use and when.
How I built it
Using EC2 DL1 instances for both of my networks across Habana accelerators I was able to train ~10 epochs at:
- < 60 s on the AttentionNet (Net 1)
- < 5 minutes for the MainNet (Net 2)
Both of my networks converge to > 0.85 accuracy.
The Habana Deep Learning Base AMI (Ubuntu 20.04) was used to pull and run their TensorFlow image. From here, I set up a development environment for my python files and began training with the htf.load_habana_module() loaded. Set-up instructions to test and train the model are included on my GitHub.
EC2 DL1
Why DL1? Why the AWS Deep Learning Challenge?
- Access to DL1 instances on Habana hardware allowed me to experiment with my model without being constrained by compute. This is probably the most important factor when developing a model that relies on being continually retrained in order to perform.
- The Habana DLAMI also provided the perfect development environment for computer vision research—making it easy to build, monitor and train the varying networks used in this project.
Datasets
Scientific datasets were obtained from UPenn, Kvasir-SEG, and the 2018 Science Bowl:
Nuclei Segmentation: https://www.kaggle.com/c/data-science-bowl-2018/overview
Brain Tumor: https://www.med.upenn.edu/cbica/brats2020/data.html
Colonoscopy: https://paperswithcode.com/dataset/kvasir
Challenges I ran into
My current development environment (laptop) is not nearly sufficient for the training required to complete this project—access to DL1 instances w/ Habana hardware allowed me to experiment without being constrained by any hardware limits
Further, multiple accelerators enabled quick re-training of the network providing the perfect environment for teaching my networks multiple tasks
Results
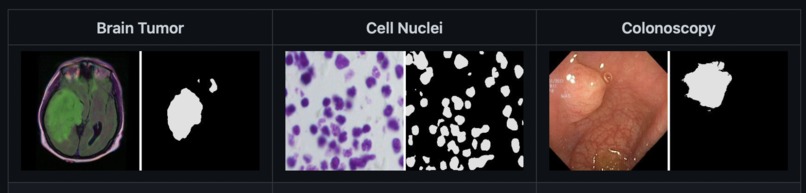
My model was able to successfully provide accurate medical image segmentation across brain tumors, cell nuclei, and colon tumors on the same neural network. This is accomplished by having my first network (CNN) tell the second one (UNet) which set of parameters it should use before passing the input image through its layers.
What's next
I want to continue working on this model by teaching it more tasks. Specifically, I would like to include diagnoses for heart disease or breast cancer screenings and then possibly even introduce it to a more clinical or academic setting for real-world patient tests.
This project focuses on technology that is especially relevant to those without access to proper healthcare; making this model public and accessible could provide accurate detection, and diagnosis of various health conditions, opening the door to new telemedicine opportunities.
Built With
- amazon-web-services
- python
- tensorflow


Log in or sign up for Devpost to join the conversation.