Inspiration
We were inspired by recent AI breakthroughs like AlphaFold that have transformed protein research, yet realized there was no single platform that made these discoveries actionable and accessible for everyone, from scientists to students. We envisioned a tool where anyone could ask a natural language question about a protein and instantly receive a full, expert-level structural and scientific analysis, with cited evidence. Driven by the desire to democratize modern molecular science and accelerate real biomedical research, we set out to build an automated, agent-based workflow that does in minutes what usually takes human experts days or weeks.
What it does
FoldForge AI is an autonomous, multi-agent AI platform for actionable protein analysis:
- Accepts natural language queries ("Find drug binding sites in p53" or "How would R273H impact EGFR?")
- Plans a stepwise workflow: extracting entities, retrieving protein structures (AlphaFold), detecting geometric binding sites, mining the latest PubMed literature, and synthesizing an evidence-backed scientific answer
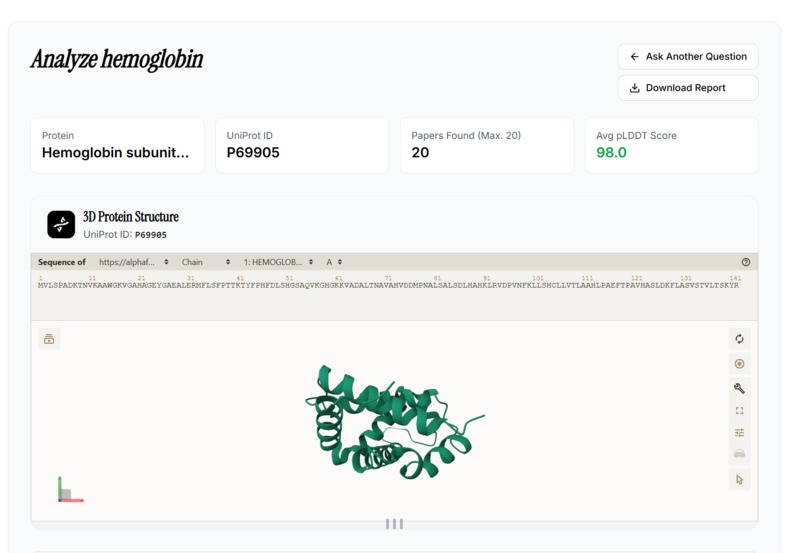
- Renders interactive 3D molecular visualizations with predicted drug pockets
- Summarizes key papers and highlights novel insights, all cited
- Outputs exportable, publication-grade reports with links, stats, and interactive elements
How we built it
- Agents: FoldForge AI orchestrates five collaborating agents: Planning (LLM), Structure (AlphaFold integration), Binding Site (geometric pocket detection in Python/Bio.PDB), Literature (NCBI PubMed + Europe PMC APIs), and Synthesis (Evidence synthesis with Nemotron LLM)
- APIs and Tools: Nemotron powers the planning and synthesis LLMs. Direct HTTP and Biopython scripts fetch real AlphaFold PDBs. Geometric detection uses adaptive neighbor-search and clustering (no ML needed). PubMed mining is robust (with retries and fallback).
- Frontend: Interactive Next.js/React dashboard allows users to launch, monitor, and explore multi-agent analysis in real time, including 3D molecular renders and step-by-step workflow progress.
Challenges we ran into
- Workflow orchestration: Getting multiple agents to pass data, handle errors, and adapt the workflow based on user query intent, without bottlenecks or silent failures.
- API reliability: Handling NCBI/EMBL downtime with seamless fallback and user-friendly messaging
- Usability: Balancing chemistry/biology complexity with a simple, fast, clear user experience, especially when surfacing confidence, caveats, and references
- Real-time UX: Streaming multi-step progress and intermediate results, so users always see where their analysis is in the pipeline
Accomplishments that we're proud of
- Our platform brings together 3D molecular data, geometric AI binding site detection, and evidence-backed scientific answers—all in real time, for anyone
- Created a system that reasons across steps, not just single queries, and takes meaningful, coordinated scientific actions
- Deployed a workflow that saves hours-to-days for tasks like lead ligand identification or mutation effect analysis, no biochemistry PhD required
- Enabled natural language, research-cited analysis for any protein question: a true step beyond Google search or static databases for scientists
What we learned
- How to integrate LLMs as both planners and answer-synthesizers in a real research pipeline, avoiding pitfalls of hallucination and factuality
- Robustness: Always provide fallback, retries, and cache for scientific APIs and model calls
- Geometric methods can be surprisingly effective (and interpretable) for binding prediction when molecular data is sparse
What's next for FoldForge AI
- Agentic Experiment Suggestions: Integrate Nemotron to propose testable lab hypotheses and related targets based on discovered structures and literature gaps.
- Deeper RAG: Add semantic retrieval over patents, datasets, or clinical trial APIs for even richer synthesis.
- Collaborative Mode: Enable shared analysis sessions and notebooks for multi-user protein research.
- Ligand/Mutagenesis Integration: Link to accessible in silico molecular docking, mutagenesis prediction, and downstream pathway modeling.
- Low-resource/edge deployment: Explore lighter models and hardware for on-site, real-time field research or diagnostics. FoldForge aims to spark a new wave of AI-accelerated, open science in protein research—putting actionable discovery in everyone’s hands.
Built With
- fastapi
- nemotron
- next.js
- nvidia
- python
- typescript


Log in or sign up for Devpost to join the conversation.