Link to Presentation Slides ☀️
Inspiration 🌎
The CDC reports that 2 billion people, or 1/4 of the world's population are infected with the Tuberculosis (TB) bacteria, with more than 10 million becoming ill with active TB each year [1]. The good news is, this is a well-researched and treatable disease. Over 90% of people survive TB given the current treatments [2]. This being said, intervention to prevent relapse can be incredibly important. TB can be detected with a chest CT scan. Researchers at Tulane University have found AI can detect and diagnose cancers in similar scans quicker and, in some cases, better than pathologists [3]. This leads us to investigate if we can create a better world for these 2 billion people affected by TB through a brighter future in the medical imaging world. Towards this, we investigate if AI can detect signs of TB relapse more efficiently and with a higher accuracy than doctors.
- https://www.cdc.gov/tb/features/wtbd/2021WTBD_Feature.html#:~:text=Two%20billion%20people%20%E2%80%93%20one%20fourth,ill%20with%20drug%2Dresistant%20TB.
- https://pubmed.ncbi.nlm.nih.gov/19584488/
- https://healthitanalytics.com/features/how-can-artificial-intelligence-change-medical-imaging#:~:text=By%20using%20AI%20in%20medical,well%20or%20better%20than%20pathologists.
What it does 🏥
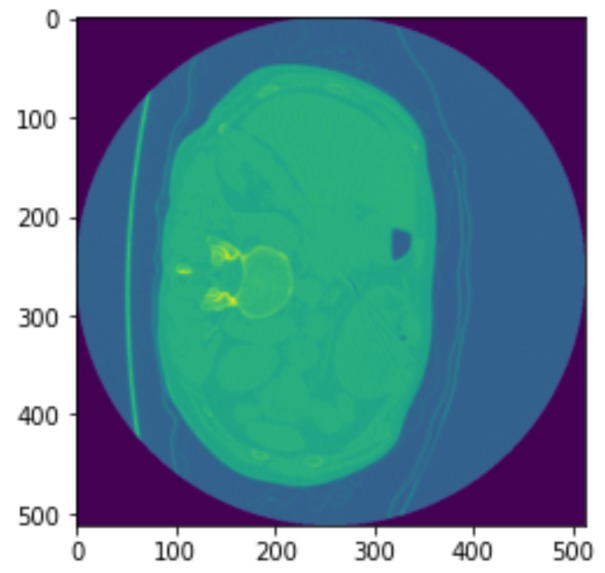
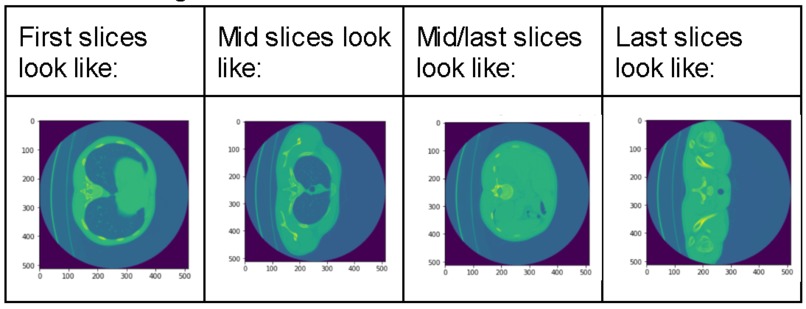
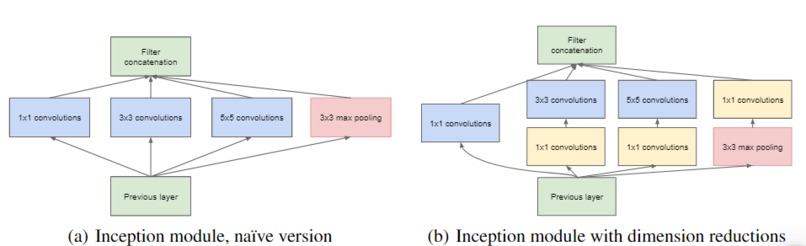
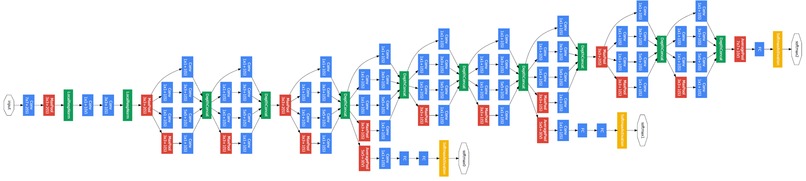
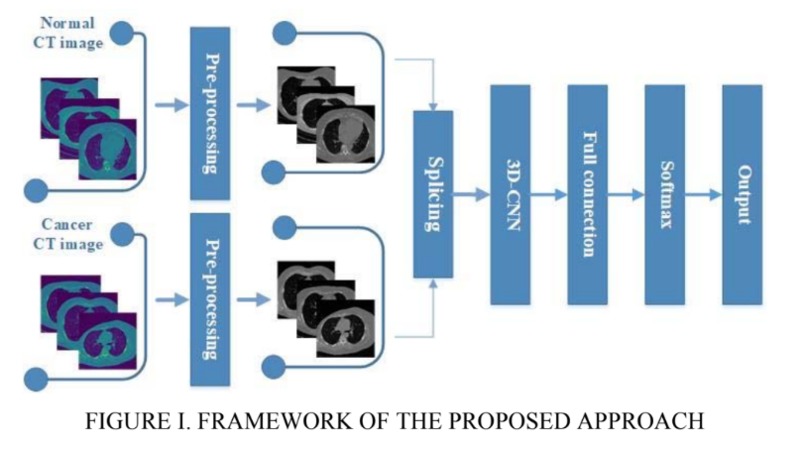
TB AI investigates the effectiveness of 2 models from the Computer Science research community in detecting key signs of TB in 3D scans of lungs. The 60 examples of 3D scans are decomposed into slices which are passed through GoogLeNet [1] and a recent 3D-CNN published in [2]. These Convolutional Neural Networks (CNNs) can then detect a binary feature present in lung caverns suggested by radiologists as a leading sign of TB relapse: presence of thick walls. This task is especially challenging as CNNs usually require anywhere from hundreds to thousands of examples to accurately make predictions.
How we built it 🧰
Our image dataset is originally in the NIfTI file format which is common for medical imaging. After running into many problems with the type of array the image was represented in the NIfTI file, we decided to convert each image to a JPG format which took a lot time and work. We then started with research into an existing GoogLeNet model, and found one on Google Colab [1]. We then modified the output layer to take in 1 class, since we purely focused on "thick walls" as our feature to classify. We did image processing by resizing the provided training image example slices, adding them to the label csv file, and using keras's ImageDataGenerator to access our data to fit our model. We then explored a more recent model, 3D-CNN [2]. We modified our NIFTI input files to fit the input size requirement for the model and debugged the model from the paper, changing the model's hyperparameters like batch size, number of training epochs, and more to achieve the best results.
- https://colab.research.google.com/github/KhuyenLE-maths/Implementation-of-GoogLeNet-on-Keras/blob/main/Implementation_of_GoogLeNet_on_Keras.ipynb#scrollTo=Zw9oBMMfgbgx
- https://github.com/AshwanthRamji/Lung-Cancer-Detection-Using-3D-Convolutional-Neural-Networks
Challenges we ran into ⛰️
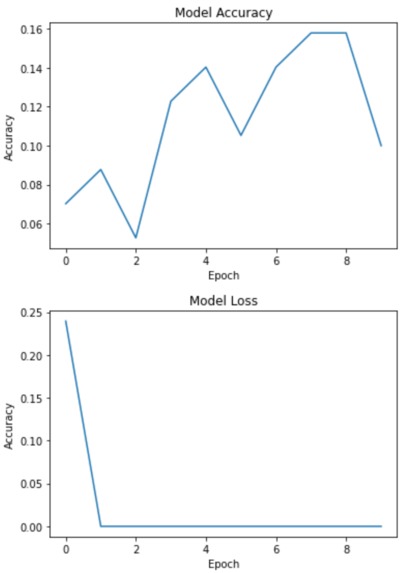
One of the main challenges we ran into was trying to implement GoogLeNet with our data. Since our training images consist of multiple slices for each lung scan, it was difficult to configure this model to take in many slices with the same labels for an image. While we haven't completely overcome this challenge, we managed to pass in one image's slices into our model and plot the accuracy. We also had to run through many trial and errors when updating the pre-existing model for our dataset. This included changing the output layer size and activation, resizing our images to fit the input layer size, and updating the y_train data to include the many slices of our example image.
Accomplishments that we're proud of 🤩
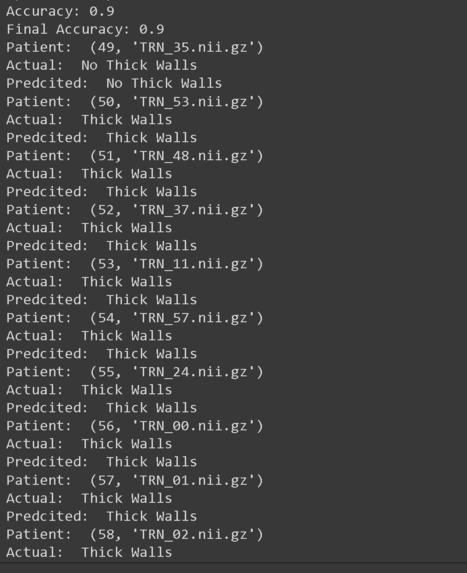
One thing we are proud of is getting the GoogLeNet model to train with our CT lung scan example. Since it gave us the most trouble, it was incredibly rewarding to get it to train without any errors. Another accomplishment is how high our accuracy is with our 3D-CNN model. Both of us have never dealt with 3D-CNNs or medical image processing, so we were happy to learn and have a working project. Last of all, we are proud that we got the chance to work on something that has a real-world application with helping others!
What we learned 💡
We both have never worked with medical image data which comes in a NIfTI file format. We learned how to navigate these files using a python module nibabel. We also learned how to implement complex CNNs with additions like inception modules, scheduled learning, and 3D convolutions. We have never applied AI to real-world issues and data, only to the classic "hello world" data like MNIST, so we learned how to take messy data in confusing formats and turn it into predictions and results that will create a better world for 2 billion people.
What's next for TB AI ⏩
The accuracy of TB AI can be even further improved. We can improve the 3D-CNN past 90% accuracy with data augmentation to produce more training examples for this few-shot learning problem. Additionally, TB AI can incorporate more visuals and images to better explain the results and demonstrate performance. Lastly, TB AI could improve it's GoogLeNet implementation by taking in a larger image dataset of multiple CT scans and their slices.
Built With
- googlecolab
- keras
- matplotlib
- nibabel
- python
- tensorflow





Log in or sign up for Devpost to join the conversation.