Inspiration
The inspiration for this project (named xrai) is to bridge the gap between AI researchers and medical professionals by creating an accessible and free prototype system that helps medical professionals understand the reality of Deep Learning tools for chest X-ray diagnostics.
What it does
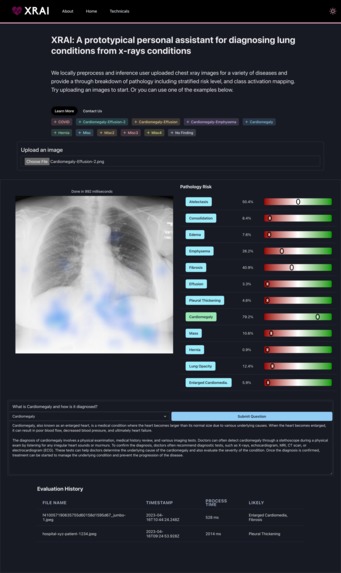
Xrai is a second opinion tool that predicts 12 different radiological findings based on data from the 7 largest public chest X-ray datasets. XRAI locally preprocesses and inferences user uploaded chest x-ray images for diseases such as Pulmonary Edema & Cystic Fibrosis, providing a thorough breakdown of pathology including stratified risk level, class activation (attention) mapping, and an LLM chatbot for further questions and learning.
XRAI takes a chest X-ray image as input and outputs the probability of each thoracic disease, alongside a class activation map (GRAD-CAM) for each pathology. The model is trained on the ChestX-ray14 dataset, which contains over 100,000 frontal view X-ray images with 14 diseases, including pneumonia, emphysema, lung opacity, and cardiomegaly. The trained model is imported into an Electron app, providing a user-friendly interface for healthcare professionals to input X-ray images and receive disease classification results.
XRAI is built for a variety of audiences, ranging from overworked radiologists who want to corroborate their diagnosis, to patients who want to learn more about their diseases, to medical students who are in training and want to supplement their education. This software could also be passively be run in bulk for healthy patients who get an x-ray done for an unrelated reason.
How we built it
For the core decision model, we used a PyTorch implementation of ChexNet through fine-tuning the 121-layer Dense Convolutional Network (DenseNet) on the ChestX-ray14 dataset, which contains over 100,000 frontal views X-ray images in 14 classes, including pneumonia, emphysema, lung opacity, and cardiomegaly. We chose to use DenseNet due to its improved flow of information/gradients through its layers, allowing for more tractable optimization. Once fine-tuned, the model was exported to tf-js through the ONNX framework to be run entirely locally on in the browser, minimizing server costs, and allowing free usage for anyone.
In order to prevent data drift, (a phenomenon that occurs when the model's knowledge slowly begins to stop reflecting data from the real world) we implemented a SSIM: a Structural Similarity Index Measure to ensure that the data our software is used for is sufficiently similar to the data it has been trained on.
Lastly we implemented GRAD-CAM: Gradient-weighted Class Activation Mapping, which is a visualization technique to understand what parts of an image a model is "looking" at in order to make its prediction. We did this by computing the gradients for each class in the tf-js model and overlaying them on top of the x-ray image as an anti-aliased heatmap.
Challenges we ran into
- Fine-tuning was difficult due to the size of the dataset.
- Developing a clean UI that is able to show all the data that we compute
- Class Activation Mapping was difficult to implement due to the details of matrix math involved and since we were doing this in javascript
- SSIM was difficult due to hyperparameter optimization
Accomplishments that we're proud of
We are proud of developing a tool that demonstrates how AI systems work and their limitations. The tool also showcases the potential of open data, which is crucial to building a public system like xrai. Additionally, we are proud of creating a tool to help teach radiology and demonstrate a model delivery system that can scale to provide free medical tools to the world.
What we learned
Building a tool like xrai requires a deep understanding of AI systems and their limitations. We also learned the importance of open data and how it can be used to build powerful and accessible tools for medical professionals.
What's next for xrai
The future of xrai involves continuing to improve the accuracy and reliability of the system while also expanding its capabilities. We also plan to make the tool more accessible to medical professionals worldwide and continue to demonstrate the potential of open data in the field of medical diagnostics.
Log in or sign up for Devpost to join the conversation.